Abstract
A DNA barcode is a DNA fragment used to identify species. For land plants, DNA fragments of plastid genome could be the primary consideration. Unfortunately, most of the plastid candidate barcodes lack species-level resolution. The identification of DNA barcodes of high resolution at species level is critical to the success of DNA barcoding in plants. We searched the available plastid genomes for the most variable regions and tested the best candidates using both a large number of tree species and seven well-sampled plant groups. Two regions of the plastid gene ycf1, ycf1a and ycf1b, were the most variable loci that were better than existing plastid candidate barcodes and can serve as a barcode of land plants. Primers were designed for the amplification of these regions and the PCR success of these primers ranged from 82.80% to 98.17%. Of 420 tree species, 357 species could be distinguished using ycf1b, which was slightly better than the combination of matK and rbcL. For the well-sampled representative plant groups, ycf1b generally performed better than any of the matK, rbcL and trnH-psbA. We concluded that ycf1a or ycf1b is the most variable plastid genome region and can serve as a core barcode of land plants.
Similar content being viewed by others
Introduction
DNA barcoding is a technique used to identify unknown materials of known species based on DNA sequences of standard genome regions (i.e. DNA barcodes)1. Before this technique can be fully utilised, a barcode that is variable enough to discriminate between species of interest and a reliable barcode reference library must be made available. The first of these requirements is more significant as it is relatively easy to build a reference library if DNA materials already exist. Ideally, a barcode should be variable enough to resolve closely related species and short enough for easy experimental manipulation and low cost. The sequences flanking the barcode should be conservative enough to facilitate the design of universal primers for high PCR and sequencing success. Presently, the candidate barcodes are selected from markers used in molecular systematics due to the limited knowledge of genome variations; we know little about mitochondrial genomes, much less nuclear genomes. Fortunately, by the end of 2013, 429 plastid genomes were sequenced and perhaps they contain a useful plant barcode.
In the past decade, several plastid genome regions such as atpF-H, matK, psbK-I, rbcL, ropC1, rpoB, trnH-psbA and trnL-F that are frequently used in plant molecular systematics have been extensively evaluated2,3,4 and the rbcL and matK genes were selected as core plant barcodes by the CBOL Plant Working Group5. Unfortunately, rbcL seems to be more suitable for barcoding lower plants than for seed plants6. One of the most highly variable regions of the plastid genome, trnH-psbA, undergoes chromosomal rearrangements (e.g., inversions and microsatellite loci) and contains a limited number of informative sites due to its short length7,8. Recently, Dong et al. found that two regions of the plastid gene ycf1 were very variable in flowering plants9.
As the second largest gene in the plastid genome, ycf1 encodes a protein of approximately 1,800 amino acids. Recent experiments showed that ycf1 is essential for plant viability and encodes Tic214, a vital component of the Arabidopsis TIC complex10. Within the plastid genome, ycf1 spans the small single copy (SSC) and the inverted repeat (IR) regions. The section of ycf1 in the IR region is short (less than one kilobase long) and conserved. In contrast, the section of ycf1 in the SSC region has high sequence variability in seed plants. This region of the ycf1 gene is more variable than matK in most taxa investigated thus far11,12 and has been used in molecular systematics at low taxonomic levels13,14,15,16,17. Two regions within ycf1, ycf1a and ycf1b, have been predicted to have the highest nucleotide diversity (π) at the species level within angiosperm plastid genomes9.
Because ycf1 is too long (5709 bp in Nicotiana tabacum) and too variable to permit the design of universal primers9, it has received little attention for DNA barcoding or molecular systematic purposes at low taxonomic levels; however, the high variability of ycf1 indicates its potential value in DNA barcoding of land plants. This paper reports the results of our evaluations of the ycf1 gene for DNA barcoding purposes. We (1) generated primers for PCR amplification of ycf1a and ycf1b and (2) tested the performance of ycf1a or ycf1b in discriminating between species compared with the plastid genes rbcL, matK and trnH-psbA.
Results
The ycf1a and ycf1b regions are the most variable regions of the ycf1 gene
According to Dong et al.9, angiosperms have two highly variable regions in the ycf1 gene, ycf1a and ycf1b. These results were verified using 136 genomes belonging to 27 genera (see the electronic supplementary material, Table S1). The exact positions of ycf1a vary slightly among plant groups, while the positions of ycf1b are quite consistent, especially in seed plants.
Primer universality
The ycf1 gene is too variable for the design of universal primers for all land plants; however, we were able to design universal primers for Bryophytes, Monilophytes, gymnosperms and angiosperms (Table 1). The ycf1mF/ycf1mR primer pair amplified the expected fragments in 32 of 34 (94.12%) Bryophyte families (see the electronic supplementary material, Table S2).
The ycf1b regions of Monilophytes are too divergent to contain satisfactory primer sites; in contrast, the sequences flanking ycf1a are relatively conserved and we designed three primers (one forward and two reverse primers, Table 1). Using these primers, ycf1a fragments were amplified from 82.80% of the samples belonging to 93 genera in 42 families (see the electronic supplementary material, Table S2).
The ycf1gF/ycf1gR primer pair was designed for conifers and cycads (see the electronic supplementary material, Table S2). The divergence of ycf1 sequences in gymnosperms is remarkable and it is difficult to identify a single pair of universal primers. Consequently, the species-rich conifers required special attention.
The ycf1bF/ycf1bR primer pair worked the best for angiosperms. The PCR success reached 98.17% samples from 219 genera in 217 families (see the electronic supplementary material, Table S2). Moreover, critical mutations at the 3′ end that would cause amplification failure were observed in some taxa. To minimise PCR failures, some substitutions to the universal primers listed in Table 1 were generated for 131 families (see the electronic supplementary material, Table S3) in the event that the universal primers failed.
Performance of ycf1 in identifying BBG woody plants
In total, we obtained 1352 sequences of matK, rbcL and ycf1 from 420 woody plant species representing 179 genera in 76 families. The sequence recoveries for rbcLb, matK and ycf1b were 99.18%, 91.43% and 85.31%, respectively (see the electronic supplementary material, Table S4). Poor PCR amplification of ycf1b was encountered for Lonicera (13 samples) and Berberis (15 samples). When samples from these two genera were excluded, the ycf1b sequence recovery reached 90.48%. Approximately 79.80% (391) of all samples contained all three sequenced fragments. Therefore, two types of datasets were generated for accurate assessments. One comprised all sequences for each marker and the other comprised the samples with all three markers. Three two-barcode combinations and one three-barcode combination were also tested for each dataset type.
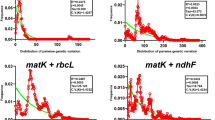
For the datasets of all sequences, ycf1b showed the highest discriminatory power of the three barcodes, discriminating 73.97% of all the species. The barcodes rbcLb (58.02%) and matK (57.56%) had similar discriminatory power (Fig. 1), much lower than ycf1b. The performance of ycf1b was slightly better than the combination of rbcLb and matK (71.31%). Combining ycf1b with either rbcLb or matK increased the discriminatory power to 81.39% and 79.83%, respectively and the use of all three candidates increased the discriminatory power to 86.33%.
To eliminate the possible effects of sample inequity on the different markers, we used datasets of 391 samples with all three markers to increase the reliability of direct comparisons of species resolution among the markers. Similar patterns were observed for these datasets. ycf1b had the highest species resolution among the three markers at 71.87%, with 54.99% resolution for matK and 55.50% resolution for rbcLb (Fig. 1).
Performance of ycf1 in identifying species within seven well-sampled plant groups
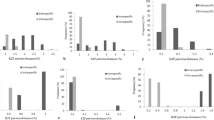
Using matK, rbcLb and trnH-psbA as controls, seven relatively well-sampled plant groups were chosen to test the discriminatory powers of ycf1b (see the electronic supplementary material, Table S5). The lengths of matK, rbcLb and ycf1b varied among the plant groups. The poly-A/T tracts longer than 9 bp were observed in the trnH-psbA sequences from six of the seven plant groups and non-homologous inversions of 2 to 21 bp were observed in five groups. The nucleotide diversity (π) of ycf1b was the highest among the four markers in the five plant groups (see the electronic supplementary material, Table S6–S12).
Using the distance method, ycf1b exhibited the highest discriminatory power among the four markers in six of the seven plant groups (Fig. 2). Combinations of matK, rbcLb and trnH-psbA did not typically increase the discriminatory rates; in contrast, the combination of ycf1b with either matK, rbcLb or trnH-psbA increased the percentage of discrimination success by varying degrees for five of the seven plant groups (Fig. 2 and see the electronic supplementary material, Fig. S1, Table S6–S12). Without ycf1b, matK and rbcLb did not contribute for either Armeniaca or Paeonia (see the electronic supplementary material, Fig. S1).
Discussion
Which gene can serve as a barcode of plants?
The question of which gene can serve as a barcode for plants remains to be answered even though the combination of matK and rbcL has been suggested as the core barcode of land plants5. Marker selection is critically important as time and money can be saved by the quick identification of a suitable barcode. Kress and Erickson had three criteria18, Ford et al. put forward five considerations19 and the CBOL Plant Working Group placed higher importance on experimental considerations. The definition of DNA barcode1 implies that the first consideration should be species resolution. The issue of a barcoding gap is not an important issue in DNA barcoding; rather, it is an important question in systematics and taxonomy. Many DNA barcoders are also systematists and the two issues have been investigated simultaneously. If a species is correctly circumscribed, the optimal barcode will show the highest probability to distinguish it from other congeners regardless of the existence of a barcoding gap. Unfortunately, it is difficult to predict when such an ideal barcode might be found. We can now attempt to identify an improved barcode while also using the old ones for some time and then eventually substitute them.
Imperfections of the existing candidates
Many candidate barcodes that cannot withstand tests of universality are only suitable for specific plant groups and are falling from common use. Only four continue to receive some use: the chloroplast genes rbcL, matK and trnH-psbA and the nuclear internal transcribed spacer (ITS). The rbcL gene was suggested as a core barcode not as a result of its power in barcoding species, but rather its historical popularity19 and possibly experimental ease. The rbcL gene has been subject to considerable criticism as a barcode for seed plants20,21,22,23; however, it may be useful for lower plants6.
Although they are advocates of matK, Ford and his colleagues acknowledged that matK had only modest performance19. The matK gene was not among the top 10 most variable species-level markers9. Although matK is more useful than rbcL6, in most cases it is not the only useful species-level barcode23,24.
The use of trnH-psbA had been well evaluated8 and although trnH-psbA is more variable than either matK or rbcL, several problems limit its widespread adoption. The extensive prevalence of inversions and insertions within species, long polystructures that cause sequencing difficulties and relatively short lengths prevent its use as a core barcode.
After a few years of disfavour, the nuclear ribosomal ITS first proposed by Kress et al.25 has again become accepted as a core barcode, as exemplified by studies using large data20,26,27,28. With the exception of ycf1, ITS has been shown to have unparalleled species resolution compared with the candidate barcodes proposed thus far; however, it suffers from incomplete concerted evolution in some cases and from experimental complexity for species of hybrid origin.
Is ycf1 good enough to be a barcode of land plants?
It is easy to identify the most variable regions in certain taxa at the species level; however, it is difficult and even unlikely, to identify such regions in all taxa. With a few isolated exceptions, the ycf1a and ycf1b regions are perhaps the most variable regions in most taxa9. A barcode should be chosen because it shows the highest species resolution in most cases rather than in specific cases. The ycf1 gene meets this criterion and can serve as a barcode of land plants.
Currently, there are two applications for DNA barcoding. One application is for flora and the other is for specific taxa. The first application is exemplified by barcoding trees in large ecological plots29,30,31,32 and a similar example was given in this study. Our test example differs from the barcoding of local flora in that more species are from the same genera, which may show reduced discriminating power. The ycf1b performed satisfactorily when compared to the core barcode combination of matK and rbcL. The second application is becoming increasingly popular for barcoding of medicines, teas and foods, etc33,34. As ycf1 was not previously identified as a potential barcode, comparisons are not currently available. In this paper, seven examples were used to show the superior performance of ycf1b compared with other barcodes for distantly related plant groups. Consequently, ycf1b is expected to be suitable for an extensive group of plants.
The ycf1 gene was slowly identified for its potential use as a barcode most likely due to its length and lack of universal primers; however, a few phylogenetic applications had been found for Pinaceae14,35, Orchidaceae13, Lamiaceae15,36 and Prunus37.
One major concern for the use of ycf1 as a barcode is the absence of ycf1 in some taxa. The ycf1 gene is functional and is not commonly lost38. It was erroneously reported to have been lost from Acorales, Poales and Passiflora; however, it is only absent from Poaceae39,40.
Experimental considerations for ycf1 use
Primer universality is an important criterion for an ideal DNA barcode. The primers for rbcLb were recently optimised6. Although matK primers had been the subject of several studies41,42,43, obtaining matK fragments from ferns and mosses continues to be a challenge. At the beginning of our work, hardly any ycf1 sequences had been deposited in GenBank; consequently, we had to generate ycf1 sequences to facilitate primer design. The amplification successes of the ycf1 primers used in this study were quite satisfactory: 98.17% for angiosperms, 90.91% for gymnosperms, 82.80% for Monilophytes and 94.12% for Bryophytes. Amplification in seed plants would not be very difficult using the universal primers. The primer performances for Monilophytes and gymnosperms were relatively poor (Table 1) due to the significant divergence within these groups. To minimise PCR failure, taxon-specific primers were generated for focal taxa (see the electronic supplementary material, Table S3).
Methods
Reconfirming the variability of the ycf1a and ycf1b regions
Very few ycf1 sequences have been deposited in GenBank. We downloaded 144 whole plastid or chloroplast genomes from GenBank (see the electronic supplementary material, Table S1). The ycf1 sequences were extracted from two or more plastid genomes from the same genera of land plants (sorted into four groups: Bryophytes, Monilophytes, gymnosperms and angiosperms), aligned using MAFFT and manually adjusted with Se-Al 2.0 as necessary. The nucleotide diversity (π) was computed using the R package with a 600 bp sliding-window and a 50 bp step size. The averages within each group represent ycf1 variability and the most variable regions were roughly identified.
Primer design
All of the ycf1 sequences deposited in GenBank were downloaded or extracted from the plastid genomes. The sequences were sorted into four groups as described above, aligned in each group using Clustal X ver. 2.0 and then adjusted using Se-Al 2.0. These sequences served as initial templates for the design of several primer pairs spanning the roughly identified regions for each group using Primer Premier 5.0 software (Premier Biosoft International, Palo Alto, CA). The successfully amplified fragments were sequenced and together with those from GenBank, served as templates for universal primer design and hypervariable region positioning.
Plant materials for primer design and discrimination power analyses
To test the universality of the primers, we used 368 samples of land plants, including 34 samples from 34 Bryophyte families, 93 samples from 42 Monilophyte families, 22 samples from 10 gymnosperm families and 219 samples from 216 angiosperm families (see the electronic supplementary material, Table S2).
Two types of tests were used to assay the performance of ycf1a, ycf1b and other markers of the same genome. The first test used almost entirely woody plant species cultivated in the Beijing Botanical Garden (BBG) of the Chinese Academy of Sciences (CAS). We used 490 samples belonging to 420 species in 76 families, including 53 samples belonging to 48 species in 5 gymnosperm families and 437 samples belonging to 372 species in 71 angiosperm families (see the electronic supplementary material, Table S4). The use of BBG materials represents a common practice of using DNA barcoding for the identification of plant materials from local flora such as from large-scale community plots.
The second test used the well-sampled groups of seed plants (see the electronic supplementary material, Table S5), namely gymnosperm Pinus (Pinaceae); basal angiosperm Calycanthaceae; monocotyledon Iris (Iridaceae); Saxifragales Paeonia sect. Moutan (Paeoniaceae); rosid Prunus sect. Armeniaca (Rosaceae); Quercus (Fagaceae); and asterid Panax (Araliaceae). We attempted to include a representative from each major angiosperm group. The species of Calycanthaceae, Paeonia sect. Moutan, Prunus sect. Armeniaca and Panax were completely sampled. Unfortunately, there are too many species in Pinus, Iris and Quercus for full inclusion in this study.
Experimental details
Genomic DNA was extracted from fresh or silica gel-dried leaves using the mCTAB method44. The 25-μL PCR reactions contained 1 × PCR buffer (with Mg2+), 0.25 mmol/L of each dNTP, 0.25 μmol/L of each primer, 1.25 U Taq polymerase and 20–30 ng DNA. The PCR program consisted of 4 min at 94°C, 34 cycles of 30 s at 94°C, 40 s at 52°C and 1 min at 72°C, followed by 10 min at 72°C. The PCR products were examined by electrophoresis on a 1% agarose gel containing ethidium bromide and visualised using an ultraviolet transilluminator. Both strands were sequenced on an ABI 3730xl DNA analyzer (Applied Biosystems, Foster City, U.S.A.) according to the manufacturer's protocols.
The primers used in this study were matK472F and matK1248R43 for angiosperm matK, Gym_F1A and Gym_R1A42 for gymnosperm matK, rbcLbF and rbcLbR for rbcLb6 and cp001F and cp001R45 for trnH-psbA.
Data analyses
The sequences were edited and assembled using a Sequencer 4.7 (Gene Codes, Ann Arbor, MI, USA), aligned using Clustal X ver2.0 and manually adjusted with Se-Al 2.0 as necessary.
BLAST, distance and tree-building methods were used to evaluate the performance of ycf1, matK, rbcL and trnH-psbA. BLAST (version 2.2.17) was used for the BBG sequences. The entire data set was used as a reference database and each sequence was used as a query. Only hits having E values < 1 × 10−5 were considered. If the hits with the highest scores included sequences from more than one species, the identification was considered to be a failure, otherwise it was considered successful following China Plant BOL Groups20.
In addition to Blast, distance and tree-building methods were used on the seven well-sampled plant groups. The Kimura 2-parameter (K2P) distances were calculated using MEGA 5.0. We considered discrimination to be successful if the minimum uncorrected interspecific K2P distance of focal species was greater than their maximum intraspecific distance. When using the tree-building method, neighbour joining (NJ) and unweighted pair group method with arithmetic mean (UPGMA) dendrograms based on K2P distances were constructed using PAUP 4.0. Species were considered to have been discriminated from one another if all of the individuals of a species formed a single and exclusive clade.
To assess the effects of multiple gene regions on the resolution of species, we compared the resolution of species as a cumulative percentage for each combination of gene regions for both the BBG samples and the seven representative plant groups.
References
Hebert, P. D. N., Cywinska, A., Ball, S. L. & DeWaard, J. R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B 270, 313–321 (2003).
Zhou, S., Renner, S. S. & Wen, J. Molecular phylogeny and intra- and intercontinental biogeography of Calycanthaceae. Mol. Phylogenet. Evol. 39, 1–15 (2006).
Jian, S. G. et al. Resolving an ancient, rapid radiation in Saxifragales. Syst. Biol. 57, 38–57 (2008).
Chase, M. W. et al. Phylogenetics of seed plants - an analysis of nucleotide-sequences from the plastid gene rbcL. Ann Mo Bot Gard. 80, 528–580 (1993).
CBOL Plant Working Group. A DNA barcode for land plants. Proc. Nat. Acad. Sci. U.S.A. 106, 12794–12797 (2009).
Dong, W. et al. Discriminating plants using the DNA barcode rbcLb: an appraisal based on a large dataset. Mol. Ecol. Resour. 14, 336–343 (2014).
Whitlock, B. A., Hale, A. M. & Groff, P. A. Intraspecific inversions pose a challenge for the trnH-psbA plant DNA barcode. PLOS ONE 5, e11533 (2010).
Pang, X. et al. Utility of the trnH-psbA intergenic spacer region and its combinations as plant DNA barcodes: a meta-analysis. PLOS ONE 7, e48833 (2012).
Dong, W., Liu, J., Yu, J., Wang, L. & Zhou, S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLOS ONE 7, e35071 (2012).
Kikuchi, S. et al. Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339, 571–574 (2013).
Oliver, M. J. et al. Chloroplast genome sequence of the moss Tortula ruralis: gene content, polymorphism and structural arrangement relative to other green plant chloroplast genomes. BMC Genomics 11, 143 (2010).
Wolf, P. G. et al. The evolution of chloroplast genes and genomes in ferns. Plant Mol. Biol. 76, 251–261 (2011).
Neubig, K. et al. Phylogenetic utility of ycf1 in orchids: a plastid gene more variable than matK. Plant Syst. Evol. 277, 75–84 (2009).
Gernandt, D. S., Hernandez-Leon, S., Salgado-Hernandez, E. & de la Rosa, J. A. P. Phylogenetic relationships of Pinus subsection Ponderosae inferred from rapidly evolving cpDNA regions. Syst. Bot. 34, 481–491 (2009).
Drew, B. T. & Sytsma, K. J. The South American radiation of Lepechinia (Lamiaceae): phylogenetics, divergence times and evolution of dioecy. Bot. J. Linn. Soc. 171, 171–190 (2013).
Majure, L. C. et al. Phylogeny of Opuntia s.s. (Cactaceae): Clade delineation, geographic origins and reticulate evolution. Am. J. Bot. 99, 847–864 (2012).
Drew, B. T. & Sytsma, K. J. Testing the monophyly and placement of Lepechinia in the Tribe Mentheae (Lamiaceae). Syst. Bot. 36, 1038–1049 (2011).
Kress, W. J. & Erickson, D. L. DNA barcodes: Genes, genomics and bioinformatics. Proc. Nat. Acad. Sci. U.S.A. 105, 2761–2762 (2008).
Ford, C. S. et al. Selection of candidate coding DNA barcoding regions for use on land plants. Bot. J. Linn. Soc. 159, 1–11 (2009).
China Plant BOL Group. et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Nat. Acad. Sci. U.S.A. 108, 19641–19646 (2011).
Zhang, C. Y. et al. Testing DNA barcoding in closely related groups of Lysimachia L. (Myrsinaceae). Mol. Ecol. Resour. 12, 98–108 (2012).
Hernandez-Leon, S., Gernandt, D. S., Perez de la Rosa, J. A. & Jardon-Barbolla, L. Phylogenetic relationships and species delimitation in Pinus section Trifoliae inferrred from plastid DNA. PLOS ONE 8, e70501 (2013).
Clement, W. L. & Donoghue, M. J. Barcoding success as a function of phylogenetic relatedness in Viburnum, a clade of woody angiosperms. BMC Evol. Biol. 12, 73 (2012).
Little, D. P., Knopf, P. & Schulz, C. DNA barcode identification of Podocarpaceae—The second largest conifer family. PLOS ONE 8, e81008 (2013).
Kress, W. J., Wurdack, K. J., Zimmer, E. A., Weigt, L. A. & Janzen, D. H. Use of DNA barcodes to identify flowering plants. Proc. Nat. Acad. Sci. U.S.A. 102, 8369–8374 (2005).
Chen, S. L. et al. Validation of the ITS2 Region as a Novel DNA Barcode for Identifying Medicinal Plant Species. PLOS ONE 5, e8613 (2010).
Yao, H. et al. Use of ITS2 Region as the Universal DNA Barcode for Plants and Animals. PLOS ONE 5, e13102 (2010).
Wang, X.-C. et al. ITS1: a DNA barcode better than ITS2 in eukaryotes? Mol. Ecol. Resour., 10.1111/1755-0998.12325 (2014).
Kress, W. J. et al. Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proc. Nat. Acad. Sci. U.S.A. 106, 18621–18626 (2009).
Gonzalez, M. A. et al. Identification of Amazonian trees with DNA barcodes. PLOS ONE 4, e7483 (2009).
Pei, N. et al. Exploring tree-habitat associations in a Chinese subtropical forest plot using a molecular phylogeny generated from DNA barcode loci. PLOS ONE 6, e21273 (2011).
Parmentier, I. et al. How effective are DNA barcodes in the identification of African rainforest trees? PLOS ONE 8, e54921 (2013).
Stoeckle, M. Y. et al. Commercial teas highlight plant DNA barcode identification successes and obstacles. Sci. Rep. 1, 42 (2011).
Little, D. P. & Jeanson, M. L. DNA barcode authentication of saw palmetto herbal dietary supplements. Sci. Rep. 3, 3518 (2013).
Parks, M., Cronn, R. & Liston, A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 7, 84 (2009).
Drew, B. T. & Sytsma, K. J. Phylogenetics, biogeography and staminal evolution in the tribe Mentheae (Lamiaceae). Am. J. Bot. 99, 933–953 (2012).
Shi, S., Li, J., Sun, J., Yu, J. & Zhou, S. Phylogeny and classification of Prunus sensu lato (Rosaceae). J. Integr. Plant Biol. 55, 1069–1079 (2013).
Wicke, S., Schneeweiss, G. M., Depamphilis, C. W., Muller, K. F. & Quandt, D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76, 273–297 (2011).
Goremykin, V. V., Holland, B., Hirsch-Ernst, K. I. & Hellwig, F. H. Analysis of Acorus calamus chloroplast genome and its phylogenetic implications. Mol. Biol. Evol. 22, 1813–1822 (2005).
Guisinger, M., Chumley, T., Kuehl, J., Boore, J. & Jansen, R. Implications of the plastid genome sequence of Typha (Typhaceae, Poales) for understanding genome evolution in Poaceae. J. Mol. Evol. 70, 149–166 (2010).
Dunning, L. T. & Savolainen, V. Broad-scale amplification of matK for DNA barcoding plants, a technical note. Bot. J. Linn. Soc. 164, 1–9 (2010).
Li, Y., Gao, L.-M., Poudel, R. C., Li, D.-Z. & Forrest, A. High universality of matK primers for barcoding gymnosperms. J. Syst. Evol. 49, 169–175 (2011).
Yu, J., Xue, J. H. & Zhou, S. L. New universal matK primers for DNA barcoding angiosperms. J. Syst. Evol. 49, 176–181 (2011).
Li, J., Wang, S., Jing, Y., Wang, L. & Zhou, S. A modified CTAB protocol for plant DNA extraction. Chin. Bull. Bot. 48, 72–78 (2013).
Dong, W., Xu, C., Cheng, T., Lin, K. & Zhou, S. Sequencing angiosperm plastid genomes made easy: A complete set of universal primers and a case study on the phylogeny of Saxifragales. Genome Biol. Evol. 5, 989–997 (2013).
Acknowledgements
This study was partly supported by grants from the Ministry of Science and Technology of China (2011FY120200, 2012BAC01B05 & 2012AA021602) and National Natural Science Foundation of China (31270239).We are grateful to Hongmei Liu, Cunming Chen, Lei Shi, Jun Wen, Ling Wang, Yi Zhong, Xu Quan, Fangfang Zhao, Ping Long, Mu Liu and Jianhua Xue for their help in field and laboratory work, and/or provision of plant materials.
Author information
Authors and Affiliations
Contributions
S.Z. and W.D. designed the study; W.D. and C.L. analyzed the data; W.D. and C.X. performed the laboratory work; S.Z., W.D., C.X., J.S., Y.Z., S.S., T.C. and J.G. collected materials; W.D., C.X. and S.Z. wrote the manuscript. All authors read and approve the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Dong, W., Xu, C., Li, C. et al. ycf1, the most promising plastid DNA barcode of land plants. Sci Rep 5, 8348 (2015). https://doi.org/10.1038/srep08348
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep08348
This article is cited by
-
Plastid genome data provide new insights into the dynamic evolution of the tribe Ampelopsideae (Vitaceae)
BMC Genomics (2024)
-
Plastid genome and its phylogenetic implications of Asiatic Spiraea (Rosaceae)
BMC Plant Biology (2024)
-
Characterization of the chloroplast genome of Gleditsia species and comparative analysis
Scientific Reports (2024)
-
The complete sequence of Lens tomentosus chloroplast genome
Acta Physiologiae Plantarum (2024)
-
Complete plastid genome structure of 13 Asian Justicia (Acanthaceae) species: comparative genomics and phylogenetic analyses
BMC Plant Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.