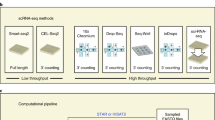
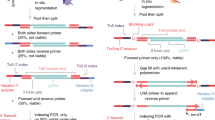
Abstract
Single-cell genome sequencing has proven valuable for the detection of somatic variation, particularly in the context of tumor evolution. Current technologies suffer from high library construction costs, which restrict the number of cells that can be assessed and thus impose limitations on the ability to measure heterogeneity within a tissue. Here, we present single-cell combinatorial indexed sequencing (SCI-seq) as a means of simultaneously generating thousands of low-pass single-cell libraries for detection of somatic copy-number variants. We constructed libraries for 16,698 single cells from a combination of cultured cell lines, primate frontal cortex tissue and two human adenocarcinomas, and obtained a detailed assessment of subclonal variation within a pancreatic tumor.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
McConnell, M.J. et al. Mosaic copy number variation in human neurons. Science 342, 632–637 (2013).
Cai, X. et al. Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the human brain. Cell Rep. 8, 1280–1289 (2014).
Knouse, K.A., Wu, J., Whittaker, C.A. & Amon, A. Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proc. Natl. Acad. Sci. USA 111, 13409–13414 (2014).
Rehen, S.K. et al. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proc. Natl. Acad. Sci. USA 98, 13361–13366 (2001).
Navin, N. et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94 (2011).
Eirew, P. et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 518, 422–426 (2015).
Gawad, C., Koh, W. & Quake, S.R. Dissecting the clonal origins of childhood acute lymphoblastic leukemia by single-cell genomics. Proc. Natl. Acad. Sci. USA 111, 17947–17952 (2014).
Gao, R. et al. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat. Genet. 48, 1119–1130 (2016).
Zong, C., Lu, S., Chapman, A.R. & Xie, X.S. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science 338, 1622–1626 (2012).
Baslan, T. et al. Optimizing sparse sequencing of single cells for highly multiplex copy number profiling. Genome Res. 25, 714–724 (2015).
Knouse, K.A., Wu, J. & Amon, A. Assessment of megabase-scale somatic copy number variation using single-cell sequencing. Genome Res. 26, 376–384 (2016).
Gawad, C., Koh, W. & Quake, S.R. Single-cell genome sequencing: current state of the science. Nat. Rev. Genet. 17, 175–188 (2016).
Adey, A. et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 11, R119 (2010).
Amini, S. et al. Haplotype-resolved whole-genome sequencing by contiguity-preserving transposition and combinatorial indexing. Nat. Genet. 46, 1343–1349 (2014).
Adey, A. et al. In vitro, long-range sequence information for de novo genome assembly via transposase contiguity. Genome Res. 24, 2041–2049 (2014).
Buenrostro, J.D., Giresi, P.G., Zaba, L.C., Chang, H.Y. & Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Cusanovich, D.A. et al. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348, 910–914 (2015).
Stergachis, A.B. et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell 154, 888–903 (2013).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Adey, A. et al. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature 500, 207–211 (2013).
Macosko, E.Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Garvin, T. et al. Interactive analysis and quality assessment of single-cell copy-number variations. Nat. Methods 12, 1058–1060 (2015).
Goryshin, I.Y., Miller, J.A., Kil, Y.V., Lanzov, V.A. & Reznikoff, W.S. Tn5/IS50 target recognition. Proc. Natl. Acad. Sci. USA 95, 10716–10721 (1998).
Olshen, A.B., Venkatraman, E.S., Lucito, R. & Wigler, M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 5, 557–572 (2004).
Ha, G. et al. Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome Res. 22, 1995–2007 (2012).
Rosenkrantz, J.L. & Carbone, L. Investigating somatic aneuploidy in the brain: why we need a new model. Chromosoma https:/dx.doi.org/10.1007/s00412-016-0615-4 (2016).
Callaway, E. 'Platinum' genome takes on disease. Nature 515, 323 (2014).
Waddell, N. et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518, 495–501 (2015).
De Kouchkovsky, I. & Abdul-Hay, M. Acute myeloid leukemia: a comprehensive review and 2016 update. Blood Cancer J. 6, e441 (2016).
Kumagai, T. et al. Epigenetic regulation and molecular characterization of C/EBPalpha in pancreatic cancer cells. Int. J. Cancer 124, 827–833 (2009).
Perkins, N.D. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat. Rev. Mol. Cell Biol. 8, 49–62 (2007).
Stahley, S.N. & Kowalczyk, A.P. Desmosomes in acquired disease. Cell Tissue Res. 360, 439–456 (2015).
Forbes, S.A. et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 43, D805–D811 (2015).
Bailey, P. et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52 (2016).
Sos, B.C. et al. Characterization of chromatin accessibility with a transposome hypersensitive sites sequencing (THS-seq) assay. Genome Biol. 17, 20 (2016).
Ramani, V. et al. Massively multiplex single-cell Hi-C. Nat. Methods http://dx.doi.org/10.1038/nmeth.4155 (2017).
Vitak, S. et al. Sequencing thousands of single-cell genomes with combinatorial indexing. Protocol Exchange https://dx.doi.org/10.1038/protex.2017.001 (2017).
Acknowledgements
The genome sequence described and used in this research was derived from a HeLa cell line. Henrietta Lacks, and the HeLa cell line that was established from her tumor cells without her knowledge or consent in 1951, have made significant contributions to scientific progress and advances in human health. We are grateful to Henrietta Lacks, now deceased, and to her surviving family members for their contributions to biomedical research. The data generated from this research were submitted to the database of Genotypes and Phenotypes (dbGaP), as a substudy under accession number phs000640. We thank the aging nonhuman primate resource at the Oregon National Primate Research Center for the banked rhesus samples, the Brenden-Colson Center for Pancreatic Care for the pancreatic ductal adenocarcinoma sample, and the Knight Tissue Bank for the rectal adenocarcinoma sample. We thank J. Shendure and Shendure laboratory members D. Cusanovich and R. Daza for helpful advice and comments, and M. Kircher for providing PCR-stage index sequences. We also thank B.J. O'Roak and R. Mulqueen for helpful discussions and manuscript suggestions. A.A. is supported by an Oregon Medical Research Foundation New Investigator Award. J.L.R. is supported by the Collins Medical Trust Foundation and Glenn/AFAR Scholarship for Research in the Biology of Aging. L. Carbone is supported by the Office of the Director/Office of Research Infrastructure Programs (OD/ORIP) of the NIH (grant no. OD011092).
Author information
Authors and Affiliations
Contributions
A.A. designed and supervised all aspects of the study. A.A., S.A.V. and K.A.T. wrote the manuscript. All authors contributed to and edited the manuscript. S.A.V. carried out all SCI-seq and GM12878 DOP library preparations, designed experiments, and performed all sequencing. A.A. and K.A.T. processed all sequence data and analyzed data. K.A.T. performed all copy-number calling. J.L.R. constructed QRP and DOP libraries on rhesus samples. A.J.F. prepared all GM12878 QRP library construction and co-prepared all SCI-seq libraries using xSDS for nucleosome depletion. M.H.W. provided tumor samples and aided in the analyses of those samples. L. Carbone supervised and provided all samples for rhesus work. F.J.S. contributed to experimental design and contributed to the manuscript. L. Christiansen produced all transposase complexes used in this study.
Corresponding author
Ethics declarations
Competing interests
F.J.S. and L. Christiansen declare competing financial interests in the form of paid employment by Illumina, Inc. One or more embodiments of one or more patents and patent applications filed by Illumina may encompass the methods, reagents, and data disclosed in this manuscript. Some work in this study is related to technology described in patent applications WO2014142850, 2014/0194324, 2010/0120098, 2011/0287435, 2013/0196860 and 2012/0208705. A.A. and S.A.V. have a provisional patent filed for some of the methods pertaining to this study.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–3, Supplementary Figures 1–25 and Supplementary Protocol (PDF 6652 kb)
Supplementary Software
Software for processing SCI-seq sequence read data. (ZIP 9 kb)
Rights and permissions
About this article
Cite this article
Vitak, S., Torkenczy, K., Rosenkrantz, J. et al. Sequencing thousands of single-cell genomes with combinatorial indexing. Nat Methods 14, 302–308 (2017). https://doi.org/10.1038/nmeth.4154
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4154
This article is cited by
-
scAbsolute: measuring single-cell ploidy and replication status
Genome Biology (2024)
-
scifi-ATAC-seq: massive-scale single-cell chromatin accessibility sequencing using combinatorial fluidic indexing
Genome Biology (2024)
-
txci-ATAC-seq: a massive-scale single-cell technique to profile chromatin accessibility
Genome Biology (2024)
-
Slide-tags enables single-nucleus barcoding for multimodal spatial genomics
Nature (2024)
-
Deep model predictive control of gene expression in thousands of single cells
Nature Communications (2024)