
RNA-seq reads mRNA, revealing which genes are active and transcribing proteins.Credit: Ella Maru Studios
A 12-year-old boy with recurrent metastatic melanoma arrived at St. Jude Children's Research Hospital for an expert consultation and additional care. The boy underwent surgery, immunotherapy and other approaches, but they failed to contain his disease, which was steadily worsening.
Coincidentally at that time, around 2017, researchers and clinicians at the hospital were collaborating on a study investigating the extent to which integrating whole-genome, whole-exome and RNA sequencing (RNA-seq) data might reveal the functional impact of genetic variants on tumour biology1. The study aimed to demonstrate the translational potential of such multiomic sequencing, and its capacity to identify previously unknown mechanisms to improve cancer treatment.
The young melanoma patient enrolled in the study, and sequencing of his tumours led to an important discovery: two genes linked to the mitogen-activated protein kinase (MEK) pathway were abnormally fused together. Faulty signalling in the MEK pathway is known to drive cancer progression in some patients, and that led clinicians at St. Jude to try a new therapeutic strategy: they treated the boy with a MEK inhibitor, trametinib, and his tumours began to shrink.
Kim Nichols, a paediatric oncologist at St. Jude, in Memphis, Tennessee, and co-director of the study, says RNA sequencing proved especially important in this case, given its ability to identify and confirm the presence of novel gene fusions that may not be readily apparent in the DNA sequence.
During the past several years, RNA-seq has emerged as a powerful technology that reveals both entirely new disease pathways and actionable genetic alterations in cancer. RNA-seq measures the presence and amounts of RNA in cells and tissues, revealing which genes are active and transcribing proteins at any given time. Along with whole-genome sequencing, proteomics, and other essential next-generation sequencing methods, it generates the sort of big data that highlight novel associations between biological endpoints. While the genome is static, RNA expression is dynamic and varied. It reflects epigenetic changes that influence how genes are expressed, independently of the DNA sequence, and provides essential clues into the biology of disease. “We rely on RNA-seq to help us classify pathogenic variants, which is hugely impactful for patient management,” says Nichols.
Upwards transcriptome trajectory
RNA-seq first emerged with next-generation sequencing in the mid-2000s. But it has evolved steadily since, along with opportunities to study where and when gene transcription occurs — both in bulk tissues and, increasingly, in single cells. At first, scientists could only sequence fresh or frozen tissues. But over the past few years, technical and bioinformatic improvements that allow researchers to assemble and extract meaningful information from RNA fragments have made it increasingly feasible to sequence paraffin-imbedded material as well. This is enabling scientists to tap into vast stores of archived biopsies and surgically resected samples in hospitals and laboratories worldwide. “These samples were never subjected to RNA-seq before because their quality was insufficient for the original methods,” says Stefan Pfister, a molecular biologist at the Hopp Children's Cancer Center in Heidelberg, Germany. “Now with good protocols for paraffin-based tissues, RNA-seq is gaining additional momentum.”
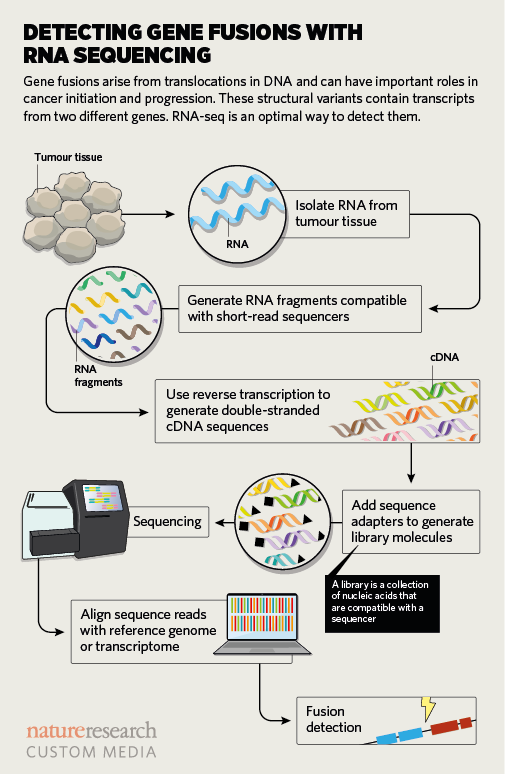
Like Nichols at St. Jude, Pfister says RNA-seq is the better technology when it comes to identifying oncogenic fusions that arise from faulty breakpoints in DNA (see ‘Detecting gene fusions with RNA sequencing’). These sorts of variants cause unrelated pieces of genes to stick together. The breakpoints can be inferred from DNA sequencing, “but they also show up as unusual RNA transcripts that you wouldn't find in a normal cell”, Pfister says. In a classic example, pieces of two unrelated genes, BCR and ABL, are fused together in nearly all patients with chronic myelogenous leukaemia.
Using RNA-seq, Pfister's team detected fusions involving a gene known as ALK in a child with aggressive high-grade glioma — a type of brain cancer with no targeted treatments. This was surprising, Pfister says, since ALK fusions had never been encountered in other brain tumours before. Working through a global consortium, the team in Heidelberg team went on to sequence glioma samples from 241 children under four years old2. They found that ALK fusions were also present in many other infants with high-grade gliomas.
ALK codes for a tyrosine kinase receptor that is also implicated in other tumour types, and several ALK inhibitors are already on the market for lung cancer. These drugs are far less toxic than the intensive chemotherapy and radiation used for treating high-grade paediatric gliomas today. “Now the paediatric neuro-oncology community is aiming to test ALK inhibitors in genetically defined subsets of children who have this disease,” Pfister says. Meanwhile, the World Health Organization (WHO) recently introduced a new diagnostic classification, Infant-type Hemispheric Glioma, for these molecularly-defined cancers3.
Defined by the transcriptome
RNA-seq is overturning other cancer classifications as well. Kenneth Aldape, a molecular neuropathologist at the National Cancer Institute's Center for Cancer Research, in Bethesda, Maryland, used the technology to investigate meningiomas, the most common adult brain cancers. Meningiomas are typically treated with surgery and have no effective pharmacologic treatments. The WHO’s histopathological classification, and the amount of tumour tissue removed by the surgeon can, to some extent, predict whether the cancer will recur. But these measures do not reliably reflect the cancer's behaviour, and some meningiomas are far more aggressive than others.
With these shortcomings in mind, Aldape and his collaborator, Gelareh Zadeh, a neurosurgeon at the University of Toronto, subjected 121 meningioma tumours to multiomic analyses: whole exome sequencing, for germline polymorphisms, somatic point mutations and somatic copy-number alterations; EPIC array profiling, for DNA methylome analysis; and RNA-seq, for transcriptome analysis.
The aim of the study, Zadeh says, was “to develop a molecular understanding of these tumours so we can help with diagnostics and, ultimately, develop new therapeutic options”. The findings moved the needle, and led to the development of a new classification scheme4. The tumours were segregated into four groups that were more predictive of clinical outcomes than the WHO’s classification scheme. RNA readouts, in particular, clustered within three of the groups, including one specific to tumour samples obtained from patients with the worst outcomes. These tumours also had high expression of histone deacetylase (HDAC), an enzyme that regulates the activity of numerous cancer-associated proteins. And in the lab, they proved susceptible to vorinostat, an HDAC inhibitor. Aldape says the RNA data provided a measure of robustness to the multiomic results. “A single platform provides useful information, but it’s also limited,” he says. “What you really want is a composite look at these tumours.”
Importantly, the four meningioma groupings were identified with unsupervised analysis, Aldape says, which means “we let the data show us what they are without imposing our preconceived notions”.
The path beyond genes
Elaine Mardis, a genomicist at Nationwide Children's Hospital, in Columbus, Ohio, describes RNA-seq as essential for discovery research in cancer, which she views as a disease not of genes, but of altered pathways. “Pathway analysis from RNA is very informative with respect to what's driving the tumour biology,” she says. “There’s so much unexplored territory in RNA, such as alternative splicing, exon-skipping events and changes in isoforms, which occur for reasons that are not apparent in the DNA sequence.” RNA-seq is especially useful for paediatric cancers, she adds, which have few gene mutations and are often driven by gene fusions or epigenetic processes.
At the same time, RNA-seq can guide scientists to where cancer mutations are located in the genome, adds Stephen Montgomery, a geneticist at Stanford Medicine, in Stanford, California. If a healthy person makes two transcripts of a particular gene, while someone with a disease makes four or five, “then we know to look around that part of the genome for the mutation that might be causing the gene to become dysregulated”, he says.
To detect these sorts of variants, however, scientists need more options to compare RNA sequences from normal and diseased tissues. “Everything is informed by our understanding of what is normal,” Mardis emphasizes. “We can't lose sight of that.” But normal comparators are in short supply, in part because the science tends to “focus on disease rather than health”, she adds. One option is to use RNA sequences taken from blood, and Montgomery has been collecting blood samples for transcriptomic analysis. But the more informative option, she says, is to compare tumour RNA with sequences extracted from adjacent non-malignant tissues, although these are rarely available.
Therefore, by necessity, cancer researchers often turn to publicly available databases for comparator sequences. One such is hosted by the Genotype-Tissue Expression (GTEx) project at the National Institutes of Health. The GTEx database contains samples collected from 54 non-diseased tissue sites across nearly 1,000 adults. Mardis says that a paediatric GTEx is in development.
Meanwhile, Nichols and her colleague, Jinghui Zhang, a computational biologist at St. Jude, continue to find translational opportunities using RNA-seq to improve patient care. Indeed, RNA data later revealed that the same fusion found in their young melanoma patient, which affects a gene called MAP3K8, is also found in roughly a third of paediatric patients with a skin cancer known as spitzoid melanoma. “RNA-seq helped in the treatment of this one patient and then opened avenues to help us understand the biology and treatment of paediatric melanoma more generally,” Nichols says.
St. Jude researchers have now begun offering whole-genome, whole-exome and RNA-seq screening to all their cancer patients. Results from their recent study of 309 patients treated at the hospital, all of them unselected for tumour type or stage, showed that 86% had variants with clinical utility1, says Nichols. “A good proportion of them were treated with targeted therapy. Some of these cancers are very hard to treat, and we now have a window of opportunity to learn a lot more about them.”


