
UQ COVID-19 vaccine project co-leader Professor Paul Young, project director Professor Trent Munro, and project co-leader Associate Professor Keith Chappell.© The University of Queensland
In late 2020, the decision was made to abandon further trials of a COVID-19 vaccine being developed by The University of Queensland (UQ) in partnership with biotech company CSL, after participants in a Phase 1 trial returned weak false-positive HIV screening results in some tests. Although there was never any chance of HIV infection, the implications for HIV testing in Australia and for the uptake of the vaccine were deemed sufficient to redirect significant Australian Government commitments to other areas, such as supplying more doses of the Oxford/AstraZeneca vaccine.
Researchers involved in the development of the vaccine, produced using UQ’s proprietary ‘molecular clamp technology’, were disappointed, but some aspects of the work gave them encouragement. “The group was devastated, as we wanted to help,” says Professor Paul Young, project co-leader along with Associate Professor Keith Chappell. “But we've taken many positives out of the project: we are further down the line in validating the platform technology and we’re very confident going forward.”
Why clamps in vaccines?
Enveloped viruses such as SARS-CoV-2, the virus responsible for COVID-19, infect a host via proteins protruding from their surface that bind to and fuse with a target cell’s membrane, allowing the virus to enter a cell. These fusion proteins are the basis of many subunit vaccines, a type in which a part of the pathogen being vaccinated against is injected to prompt the body to efficiently raise an immune response that can protect against the live virus.
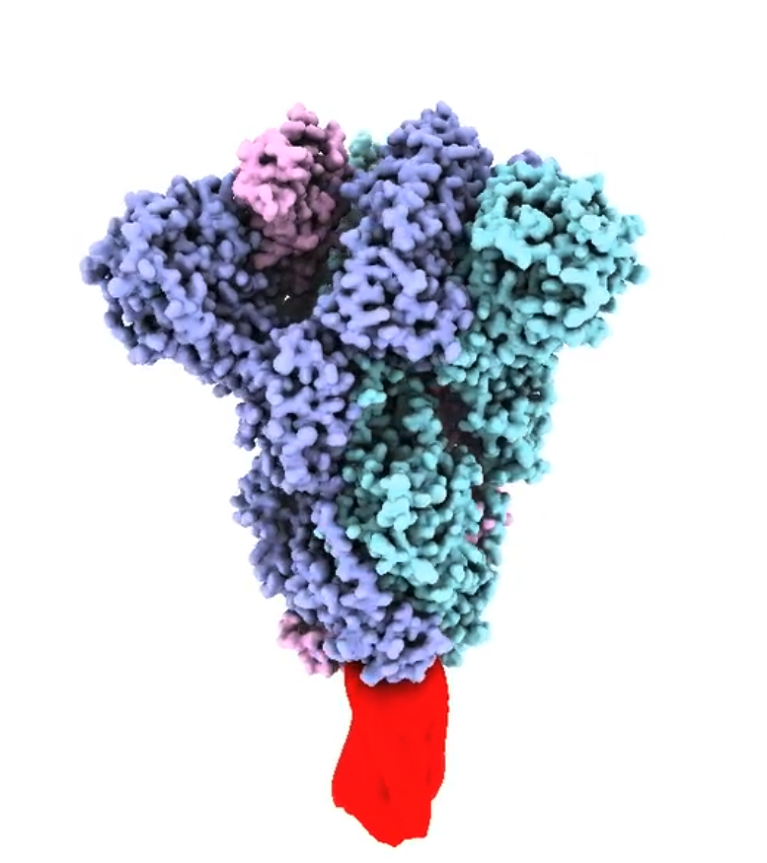
Structural model of the trimeric SARS-CoV-2 spike protein ectodomain (prepared by D Watterson), stabilized by the molecular clamp (red). https://vimeo.com/392914003© Dr Dan Watterson, The University of Queensland
The SARS-CoV-2 spike or fusion protein is a trimer: a group of three units, each consisting of a receptor-binding subunit, S1, and a subunit responsible for fusion, S2. The three S1 and S2 subunits form a fusion spike embedded in the viral membrane and held in a very particular conformation until it binds to a target cell.
“The fusion protein on the surface of the virus behaves remarkably like a mousetrap in that it's held in a highly tensed form,” explains Young. Binding to a receptor triggers a very rapid structural change, with some parts of the protein flipping through 180°, pulling the viral membrane and the host membrane close together to allow them to fuse.
“The trouble is that the final stable post-fusion form of the protein is not what is on the viral surface and so is not an appropriate form to stimulate a strong protective immune response in a vaccine. Many of the important antigenic sites on the spike protein become hidden or disappear because they literally change shape.” The same conformational change occurs when the spike protein is isolated or made by recombinant technology to produce a soluble subunit vaccine.
UQ’s molecular clamp technology was designed to hold the spike protein in its original form to produce a useful subunit vaccine.
Trying to quickly clamp down on COVID-19
Between 2008 and 2011, Keith Chappell and others had been involved in a study on respiratory syncytial virus-induced disease. That study showed that in naturally infected individuals the majority of potent neutralizing antibodies were those responding to the pre-fusion form of the protein, not the post-fusion form. This was the impetus for building the molecular infrastructure to maintain a spike protein in its pre-fusion form.
The UQ clamp that was developed consists of fragments of the fusion subunit of HIV’s spike protein, gp41. While other trimerization proteins, such as foldon from the T4-phage fibritin, were tried, the resulting structures were unstable and wouldn’t keep well in storage. Further structural modification was needed to achieve stability, whereas fusion proteins clamped using the gp41 fragments were stable on their own.
The gp41 fragment sequences, which are shaped as coils known as alpha helices, link to the end of each of the spike protein’s three fusion subunits (S2s). The alpha helices spontaneously fold and interact with helices on the other monomers. “What you end up with is a very stable, tightly bound six-helical bundle comprising three sets of two alpha helices folded back on themselves," explains Chappell. The bundle structure stabilizes the spike protein in its pre-fusion conformation.
Previous work had identified three main areas of the trimeric spike proteins of many enveloped viruses like SARS-CoV-2 where modification might improve stabilization and expression. Using the genetic sequence of the virus that produces the SARS-CoV-2 spike protein, the researchers used a trial-and-error approach to generate more than 200 versions of the spike protein in a short period. These were monitored to identify those most efficient at generating a stable spike trimer and most efficiently expressed in cells. “This required no knowledge of the protein structure,” stresses Young, “which can significantly speed up the process of producing vaccines for newly identified viruses.”
The UQ group had previously generated clamped fusion proteins of more than 10 different viruses using gp41, including Ebola, MERS, a wide range of influenza types, herpes simplex, and Nipah viruses. An important proof-of-principle result was rapid antigen production against two, somewhat exotic, but potential emerging viral pathogens - a paramyxovirus first identified in Achimota in Ghana and a mammarenavirus first identified in Wenzhou in China.
“We asked whether we could take a sequence from these two viruses, apply the principles we’ve learnt and make a vaccine candidate very quickly, without knowing anything about the protein structure," explains Chappell. “The whole intent was to apply the technology to the threat of emerging disease,” notes Young.

UQ COVID-19 vaccine team member Eve Radunz.© The University of Queensland
False positives stop planned trials
The detection of false HIV positives in all of the trial participants, albeit at very low levels and only in some assay types, halted progress to the next stage of trials. “The design of the clamp was such that the major antibody binding sites of gp41 weren’t present, so we had anticipated that if there was an antibody response to HIV it would be very minor,” says Young. HIV diagnostic assays only detect human antibodies, so it was not until the Phase 1 trial that researchers could really test for the impact of diagnostic interference. Participants were advised ahead of the trial of this possibility.
“The diagnostic cross-reactivity was very weak, and although there was never any chance of HIV infection, a wide-scale rollout of the vaccine would have a real impact on the way we diagnose HIV,” says Young. The association with HIV may also have had an impact on vaccine uptake. While their Phase 1 trial will be completed this year, the planned Phase 2/3 trials have been taken off the table.
“You need to remember we were in the middle of a research project, and the plan was to look at a wide range of clamp constructs, but when the pandemic hit we went with using what we had in our toolbox, the gp41 construct, which we knew was working well in animal models. If COVID-19 had hit in 2021 and not 2020 we would have had an extra year to do that work,” says Young.
Current work is focused on looking at similar clamp constructs from other viruses and systems that have the same level of stability as gp41 in order to re-engineer a clamp 2.0.
“I don’t use the word ‘failure’,” says Young. “It is the lessons learnt from these experiences that help you go forward.”



 Sorghum research heats up
Sorghum research heats up
 Spider venom shows potential as treatment for epilepsy, stroke and pain
Spider venom shows potential as treatment for epilepsy, stroke and pain
 Solar-driven industry: powering the future
Solar-driven industry: powering the future
 Superbugs: Finding the path of least resistance
Superbugs: Finding the path of least resistance
 Switching off chronic inflammation
Switching off chronic inflammation
 Reprogramming autoimmune diseases
Reprogramming autoimmune diseases
 Directing resources where reefs need them
Directing resources where reefs need them
 Molecular clamp vaccines: lessons from a setback
Molecular clamp vaccines: lessons from a setback
 Could microbes make barren ore waste productive?
Could microbes make barren ore waste productive?