The microbiota in the human gut is the source of our 'second genome', and sequencing is revealing its secrets.Credit: ChrisChrisW / iStock / Getty Images
Microbiome research is changing how we see ourselves as humans. The body harbours at least as many microbial cells as human cells, and our microbial gene catalogue has more than a million genes. Yet, we still know very little about the function of much of the ‘second genome’ and how it influences health.
“The gut microbiome is like a wild-card organ in the human body,” says Gary Schroth, Distinguished Scientist and VP at sequencing technology developer, Illumina, based in San Diego. “Everybody has a different gut microbiome that has a huge effect on how we digest food and metabolize drugs.”
Large collaborative projects, such as the Human Microbiome Project in the US and MetaHIT (Metagenomics of the Human Intestinal Tract) in Europe, have produced a wealth of data on the gut’s microbial communities in people with specific diseases, as well as in healthy populations. These projects revealed the microbial diversity among individuals, and highlighted the need for deeper analysis of the microbiome to understand its contribution.
Perturbation of the gut microbiome is associated with numerous diseases — and not just those that affect the digestive system, such as inflammatory bowel disease (IBD). Heritable immune-mediated diseases such as asthma, neurological conditions including autism, and genetically driven diseases such as cancer, have all been linked to dysregulation of the gut microbiota. Translating microbiome research into treatments that modulate the microbiome could transform the management of common diseases.
This is an opportunity that Ami S. Bhatt, a physician-scientist at Stanford University in California, finds very attractive. “We have known for decades that specific human gene mutations are associated with disease, but fixing those are really difficult,” she says. ”However, there are also microbiome compositions associated with disease, and we already have interesting tools to alter them.”
Bhatt’s research focuses on the communication, not just between microbes and the human cells, but also within microbial communities. “We are trying to understand what microbes are there, what they encode, what they produce as communication signals and how these change over time,” she explains. Her team is investigating how manipulating these signals could improve health and patient outcomes.
Technologies to mine the microbiome
For decades, sequence-based bacterial analysis relied on genes encoding the 16S ribosomal RNA (rRNA). 16S rRNA genes consist of highly conserved nucleotide sequences separated by variable regions that are genus- or species-specific. Researchers find the conserved regions, then amplify and analyse the interspersed variable regions — comparing them with reference sequences to identify the bacterial species present.
However, because only one gene out of each genome is examined, 16S sequencing provides very limited information about the functional potential of a microbial community. It also underestimates the total diversity of microbes in a sample, as it can’t easily distinguish bacteria that are closely related and it doesn’t detect other microbes, such as fungi and viruses, which also contribute to microbiome function.
The falling cost of sequencing and the development of improved computational methods has led to the widespread adoption of ‘shotgun metagenomics’. Using this approach, researchers can analyse the entire genomic content of a sample by sequencing extracted DNA fragments and either comparing them to reference databases or assembling them based on overlapping sequence similarity. With shotgun metagenomics, researchers can identify genes or gene fragments and predict the function of their encoded proteins. It is a powerful technique that provides information on the functional potential of a microbial community, but since sequence reads are not confined to a single locus, such as the 16S rRNA gene, it requires a greater depth of sequencing to achieve comparable detection rates. This can make shotgun metagenomics prohibitively expensive for large-scale studies.
Nevertheless, researchers are increasingly switching to shotgun metagenomics. “Compared to 16S sequencing, shotgun metagenomics provides much richer information,” says Schroth. And, he adds, costs can be contained without compromising results by adjusting the depth of sequencing.
Shallow-depth sequencing relies on microbial reference genomes to reassemble the fragments, which limits its utility. For the vast majority of microorganisms, these do not yet exist. “We work largely in a reference-free space. We can’t map a lot of the reads that we generate to reference genomes,” says Bhatt. “Coming up with ways to generate novel genomes is an exciting challenge”, and one that can benefit from deep whole-metagenome shotgun sequencing1.
Functional bioprospecting
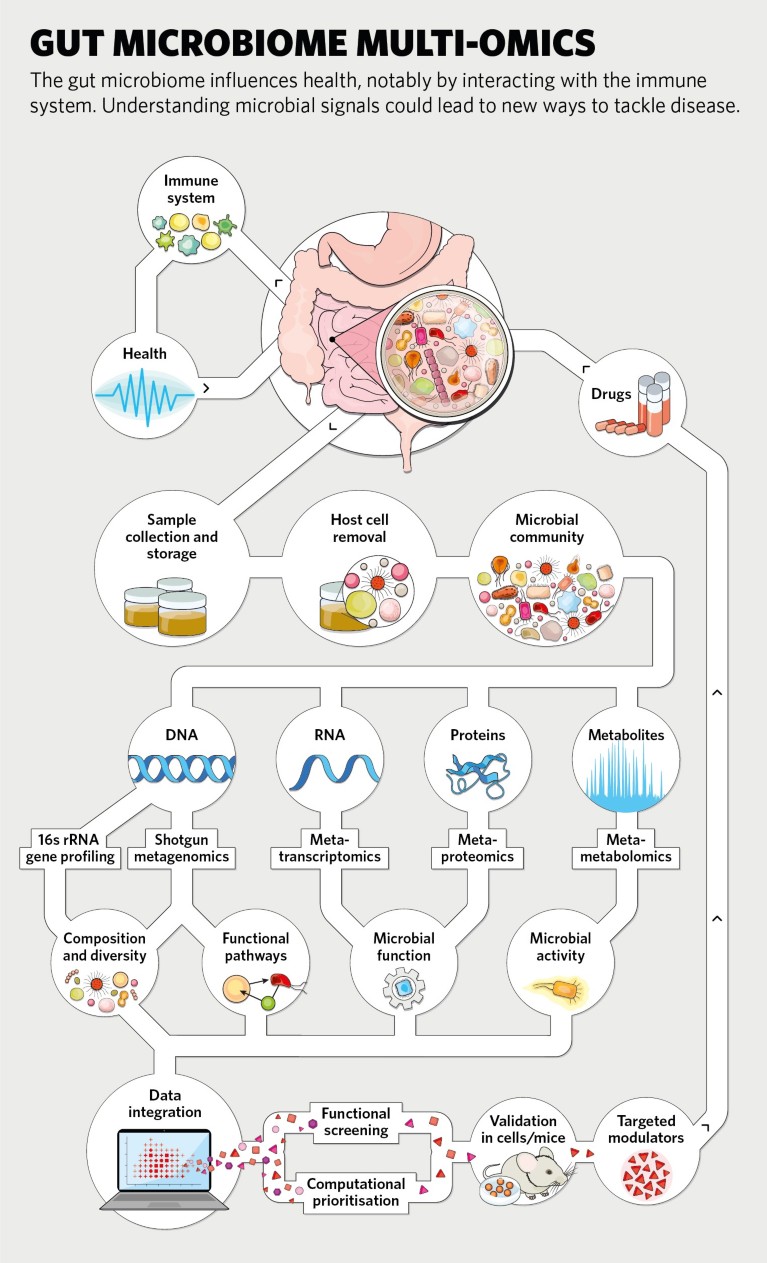
Ultimately, to understand the function of microbial communities in the gut microbiome, it is necessary to determine which genes are expressed and being translated into proteins. Thus metagenomic data is increasingly being complemented by RNA sequencing data that allows metatranscriptomic analyses, and molecular separation methods, coupled with mass spectrometry or nuclear magnetic resonance to build metaproteomic and meta-metabolomic profiles of the constituent communities.
“More researchers are using shotgun metagenomics alone or in combination with one other approach such as community metabolomics,” says Curtis Huttenhower, a computational microbiologist at the Harvard T.H. Chan School of Public Health in Boston. “Linking sequencing with high-throughput molecular tools allows us to ‘see’ microbial communities in new ways.” Meta-metabolomic analyses can help to validate metagenomic findings by identifying mechanisms linking gut microbial metabolism and cardiovascular disease2 risk or insulin sensitivity3, for example.
Huttenhower is also a principal investigator of the Human Microbiome Bioactives Resource (HMBR), which provides platforms and methods to integrate 16S sequencing, shotgun metagenomics, metatranscriptomics and meta-metabolomics for microbiome discovery. The HMBR’s goal is to identify microbial products that are important for human disease and to determine their functions. “We are working our way through a shockingly large number of uncharacterized genes, gene products and metabolites that are involved in disease phenotypes and modulated by immune activity or diet,” he says. Indeed, more than 50% of the genes in the human gut microbiome have not been characterized4. “It will take us a while to get there,” he concedes.
Learning to manipulate the gut microbiome
In the last decade, clinicians have been able to modify their patients’ microbial ecologies, by replacing a ‘bad’ microbiome with a ‘good’ one through a faecal transplant. These seem to work for some diseases, particularly recurrent Clostridium difficile infection, but there are still concerns about its efficacy for other gastrointestinal conditions5.
Away from gastrointestinal conditions, the situation is even murkier about the role of the microbiome and how it might be influenced. Certain gut bacteria are associated with post-transplant infection in cancer patients who have received a hematopoietic stem cell transplant6,7. “In around 40% of patients receiving stem cell transplantation that develop blood stream infections, we can trace the pathogen causing infection to the gut microbiome,” says Bhatt.
Furthermore, the incidence and severity of graft-versus-host disease, when immune cells from a donor transplant attack the recipient’s tissues, have been linked to gut microbiome changes, including low microbial diversity8. Having a diverse ecosystem of gut microbe types can help to modulate host inflammation and immune tolerance. Bhatt’s team are currently examining how faecal transplants and prebiotics can change the microbiome composition of people undergoing stem cell transplants and improve their outcomes.
There may be other treatment options that don’t involve replacing the whole microbiome. Last year, Bhatt and colleagues reported the existence of tens of thousands of previously overlooked small proteins (shorter than 50 amino acids in length) that are likely to have important roles in microbial communication, both with other microbes and with host cells9. Further understanding of the roles of these proteins could lead to more targeted and potentially more effective therapeutic approaches.
Huttenhower’s team is also interested in targeting specific components within the gut ecology, either in human cells or in microbes, which requires detailed functional understanding. “The hope is that we can design small molecules that target just the right host immune receptor or that inhibit or activate just the right microbial metabolic pathway to fix a particular problem,” he says.
As part of their work with the HMBR on IBD, which includes Crohn’s disease and ulcerative colitis, they have been able to computationally predict a role for previously uncharacterized bile acids and microbial proteins in gut inflammation. By using microbial and mammalian cell screens to test mechanisms of action, and gnotobiotic mice, in which all the microorganisms are either known or excluded, they can confirm predicted bioactivities in controlled experimental settings and assess the contribution of these bioactive compounds to IBD pathogenesis10.
Outlook and challenges
To improve the functional analysis of the gut microbiome and develop targeted approaches to modulate it, Bhatt emphasizes the need to collect and sequence samples at different timepoints. “Microbes are not stable or stationary, they are living organisms that can have a selective advantage in different clinical settings,” she says. She also looks forward to further technological developments to help examine more accurately how microbial genomes within a community change over time.
Current methods are also poor when it comes to detecting the exchange of genes between microorganisms, known as horizontal gene transfers, which are a critical part of the way that microbes adapt to different circumstances, such as the presence of antibiotic for example. Bhatt’s team are exploring ways to overcome this detection issue by sequencing longer DNA fragments that can be aligned more easily, leading to more accurate bacterial genome assembly.
There is little doubt that NGS technologies have transformed the field of metagenomics. Further advances in analytical methods, including machine learning, to integrate multi-omic microbiome data will have a profound impact on human disease. “As the number of tools for detecting and assaying microbes has grown,” says Huttenhower, “so has the diversity of questions that can be asked, and the number of potential translational applications.”


