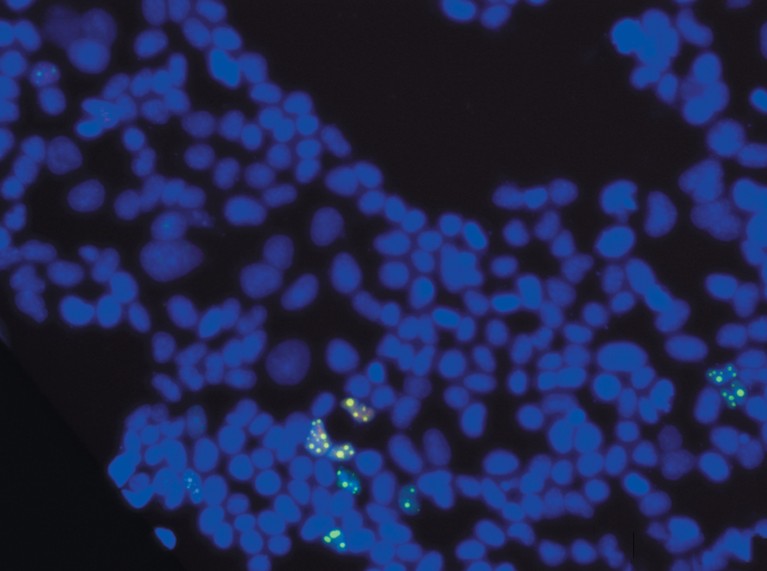
The green speckles show pentatricopeptide repeat (PPR) bound to a particular sequence of RNA. The expression plasmids containing GFP-rPPR, which binds to NEAT1 RNA, were transfected into a HEK293T cell, a derivative of human embryonic kidney cells, and observed via fluorescence microscopy following Hoechst staining.
What started as a quest to understand the genetic features of organelles inside a weedy plant, could end up becoming the next revolutionary advance in gene-editing technology.
It might not roll off the tongue as easily as CRISPR, but PPR is poised to become the next big thing in gene-editing technology, and unlike most CRISPR systems, which only target DNA, PPR-based tools allow the precise manipulation of RNA as well.
Short for pentatricopeptide repeat, PPR is a type of sequence motif found in the proteins of many plants. These proteins ordinarily help control gene activity inside the subcellular power stations, mitochondria and chloroplasts. But over the past eight years or so, researchers have discovered how to use PPR proteins for therapeutic gain.
“RNA editing is very powerful technologically,” says Takahiro Nakamura, a plant molecular biologist at Kyushu University in Fukuoka, Japan, who also serves as the chief scientific officer at EditForce, a leading developer of PPR genome-editing tools. Noncoding RNAs have been linked to the majority of diseases caused by dysregulation of gene activity and RNA splicing defects are thought to play a role in roughly 15 per cent of all human disease.
PPR platforms, says Nakamura, could help unlock the therapeutic potential of RNA editing for diseases ranging from age-related macular degeneration (AMD) to various neurological conditions.
Moreover, says Nakamura, their technique is “mature, the efficacy is very high, and it can be applied to target any RNA of interest.”
In human cells and mouse models, the researchers have shown they can design PPR proteins that alter the splicing of regulatory RNAs involved in eye and nervous system diseases. The researchers have also adapted the platform to steer the localization of RNA transcripts inside cells, as well as to boost the efficiency by which the cell’s protein-production machinery reads the genetic instructions encoded in RNA.
Through EditForce, Nakamura, and his cofounder, Yusuke Yagi, are now improving the technology. Meanwhile, they are moving forward with several drug development programmes aimed at addressing needs in oncology, ophthalmology and neurology.
Grew from the weeds
Nakamura began studying PPR proteins in the early 2000s, soon after their initial discovery by two independent research teams in France. Working as a postdoctoral fellow in Israel, he helped characterize a protein from the weedy thale cress plant that contained thousands of PPR motifs, each about 35 amino acids long, and each needed to maintain the protein’s full RNA-binding capacity in the chloroplast.
At Kyushu, Nakamura continued to examine this same protein, homing in on which particular amino acids underpin specificity and affinity for target RNA. And in 2013, he and Yagi finally deciphered the molecular code by which PPR proteins recognize particular RNA sequences — a finding that opened up the possibility of creating custom proteins tailored to bind and manipulate any RNA in a living cell.

Dr. Takashi Ono is president of EditForce.
After adapting the technique to allow for DNA modification, and patenting all the underlying intellectual property (IP), the researchers formed EditForce in 2015, with industry veteran, Takashi Ono, serving as the company’s president and chief executive. Ono and his team now hope to establish EditForce as a major player in the global genome-editing market, one that analysts forecast could be worth as much as US$10 billion by 2025.
Working with an undisclosed startup, EditForce is already using its DNA-binding PPR technology to build more potent engineered T cell therapies for cancer patients. And with a Nagoya-based pharma company called Japan Innovative Therapeutics, EditForce is designing RNA-targeted proteins aimed at correcting imbalances in gene expression that contribute to AMD.
Another pharma partnership focuses on suppressing the activity of RNA molecules linked to neurodegenerative disease. Plus, the company’s pipeline includes many early-stage drug discovery projects intended to manipulate all manner of RNA biology. “By targeting specific RNAs,” Nakamura explains, “we have developed various applications for the technology — including translational enhancement, splicing control, change of localization, and in vivo visualization of RNA activity inside cells.”
IP advantage
For now, EditForce’s profile remains somewhat under the radar, and the companies focused on CRISPR gene editing continue to gain the most media attention — although not always for the best reasons. Several CRISPR companies have netted multi-million dollar co-development deals with large pharmaceutical firms, but they have also racked up legal bills in patent disputes over who owns rights to the technology platform.
EditForce has no such problems. “Our free-standing IP is a big advantage,” says Nakamura. The company has the freedom to operate, without fear of having one day to pay hefty licensing fees to anyone else. And with the unique ability of PPR proteins to manipulate both DNA and RNA, Nakamura thinks the system has the “highest engineering potential” among all technologies in the gene-editing toolkit.
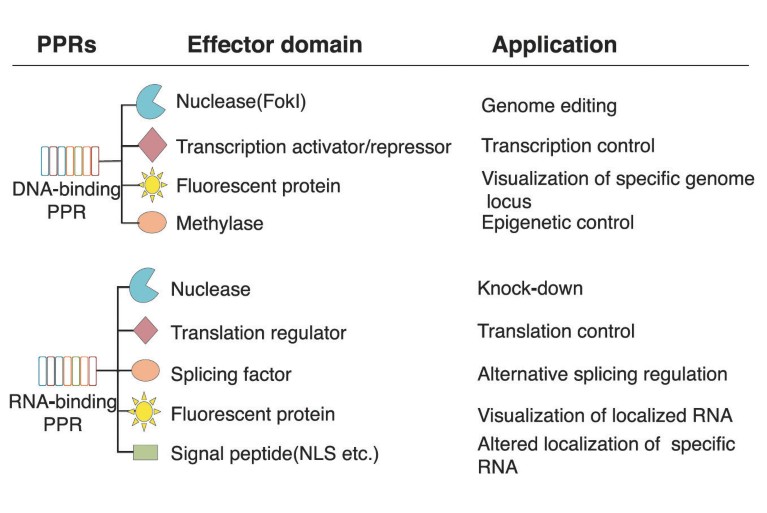
A graphic outlines the potential applications of both RNA- and DNA-binding pentatricopeptide repeat (PPR).


