
A refashioned CRISPR-Cas9 system called Target-AID doesn’t need to cleave DNA’s double-stranded structure to change a gene’s function. Instead, it performs highly targeted mutagenesis, greatly increasing the chances for microbes, such as gut bacteria, to survive gene editing. Japanese biotechnology company, Bio Palette, will harness this new technology to use modified microbes to improve people’s health.
Target-AID was developed at Kobe University in 2016 by Professor Keiji Nishida and Professor Akihiko Kondo. Derived from primitive marine animals called sea lampreys, an activation-induced cytidine deaminase ortholog, PmCDA1, was engineered to form a synthetic complex with a nuclease-deficient CRISPR-Cas9 for delivery to DNA, forming Target-AID.The tool’s mutagenesis mechanism changes the possibilities for gene-edited microbes, says Nishida, who, along with Kondo, is a director at Bio Palette. Current technologies that cleave DNA induce too much cytotoxicity to be useful for most microbe editing, he explains. “The original CRISPR-Cas9 killed almost 100% of bacteria very efficiently,” he says. “But in our version, we see almost no toxicity in, for example, yeast cells, and only a small amount of toxicity in E. coli, which is not going to be a problem at all.”
Microbiome therapeutics
Modified microbes should be useful to microbiome therapeutics. Scientists are increasingly linking the microbiome — the 100 trillion microbes in the human body — to health. However, it has proven difficult to permanently change the proportion of different microbes in the microbiome, says Nishida. Even just trying to increase good microbes or decrease bad microbes is hard.
Microbes often don’t survive gene editing, and while people can try to change their microbiome through dietary choices, specialized diets are hard to maintain and the results are often mixed.
“Our base-editing technology has the potential to solve this issue,” explains Nishida. “It’s possible that we can contribute to strengthening the breeding of good microbes and to weaken bad microbes through specific gene disruption, without changing the population, or even to breeding microbes with stronger colonization abilities.”
This could help sidestep the use of recombinant microbes — microbes in which new sections of DNA have been inserted. In the future, these microbes may face regulatory issues, notes Nishida.
Bio Palette is also working with clinicians on how to apply these techniques to treat a range of diseases, including inflammatory bowel disease, arteriosclerosis, diabetes, colorectal cancer, dermatitis, and periodontitis.

Target-AID and Target-G were developed by Keiji Nishida and Akihiko Kondo.
Pinpoint mutagenesis
One of Target-AID’s advantages is that it’s very precise, which means that it can disrupt gene function and add new functions, with highly predictable results.
Target-AID can induce base substitution in a very specific pattern and in a very narrow region — fewer than five bases — and offers the ability to edit several areas simultaneously, with a low risk of instability.
This accuracy is partly because Target-AID doesn’t need to cleave DNA to edit bases. In ordinary genome editing, cutting DNA’s double strand can induce insertions or deletions in the break, either during the repair process or when inserting sequences through homologous recombination.
A very different strategy is employed by Target-G, another proprietary base-editing technology developed at Kobe University. Target-G induces random base substitutions within a region of roughly 1,000 base pairs, using glycosylase as the base-converting enzyme. Also licensed by Bio Palette, the Target-G technology’s wider mutational range will be useful for semi-rational evolutionary engineering, microbe breeding, or library development for the discovery of useful traits.
A global shift
Recently, microbiome therapeutics has become a busy investment space. In 2017, total global venture investment exceeded US$500 million. In recognition of the field’s rapid expansion, in 2018 the US Food and Drug Administration pointed to a growing need to build stronger regulatory frameworks around live microbiome-based products and therapies.
It’s no surprise then that Bio Palette isn’t the only company to see the promise of base editing techniques. Beam Therapeutics Inc., a US-based biotechnology company, has licensed similar base-editing intellectual property from Harvard University, the Broad Institute, Massachusetts Institute of Technology, and Editas Medicine, in order to develop precision genetic medicines.
In March 2019, the two businesses signed a cross-licence agreement giving the other access to relevant intellectual property. The agreement also gave Bio Palette exclusive rights to focus on microbiome-related therapeutics in Asia. In return, Bio Palette licenced their base-editing intellectual property to Beam for use in all other human therapeutics. However, if Beam decides not to develop microbiome therapeutics outside Asia, Bio Palette will have the right to pursue this opportunity.
Making use of modified bacteria for better microbiome therapeutics is a highly anticipated development. Bio Palette is poised to be at the forefront of this push with key proprietary technology and a developed intellectual property strategy. The company is currently open to new partnerships, including with pharmaceutical or food/nutrition companies interested in microbiome therapy-related R&D collaborations.
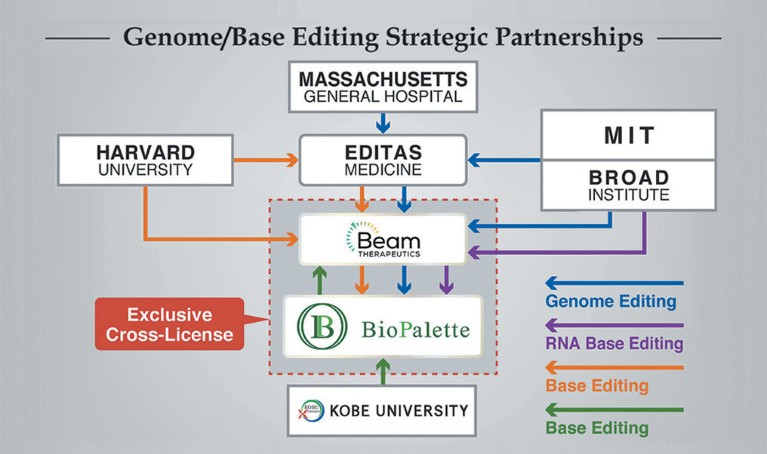
Bio Palette and Beam Therapeutics signed a cross-licence agreement in 2019.


