Tomorrow’s AI applications will not happen without research being shared openly in repositories such as that maintained by the European Molecular Biology Laboratory’s European Bioinformatics Institute near Cambridge, UK.Credit: Edmund Sumner/View Pictures/Universal Images Group/Getty
“I wake up and type AlphaFold into Twitter.”
John Jumper couldn’t hold back his excitement. He was talking to Nature in April for a News Feature on how software that can predict the 3D shape of proteins from their genetic sequence is changing biology (Nature 604, 234–238; 2022). Jumper leads the team at London-based company DeepMind that developed the AlphaFold software. Last week, DeepMind, part of the Google family, announced that its researchers have used AlphaFold to predict the structure of 214 million proteins from more than one million species — essentially all known protein-coding sequences.
‘The entire protein universe’: AI predicts shape of nearly every known protein
AlphaFold is clearly one of the most exciting developments to hit the life sciences in recent decades. As of last week, more than 500,000 researchers from 190 countries had accessed more than 2 million protein structures that DeepMind had released since last July. The structures are available in an open database jointly maintained with the European Molecular Biology Laboratory’s European Bioinformatics Institute (EMBL-EBI) near Cambridge, UK — an intergovernmental organization committed to sustaining biological data as a public good. Already, the database has been mentioned in more than 1,000 research papers.
Artificial intelligence (AI) is in the life sciences to stay. But to validate and build on insights arising from this technology, research organizations need to establish close working relationships between theoretical, experimental and computational disciplines.
Moreover, companies other than DeepMind need to seize this opportunity and commit to working with open repositories such as those maintained by EMBL-EBI. Their data, and their software needs to be freely shared — enabling development of the next generation of AI tools.
Over the past year, scientists have applied AlphaFold in all sorts of ways. Some have used its predictions to identify new families of proteins (which now need to be verified experimentally). Some are using it to help the search for drugs to treat neglected diseases. Others have looked at genetic sequences gathered from ocean and wastewater samples. The intention here is to identify enzymes whose predicted structure suggests that they have the potential to degrade plastic.
Artificial intelligence in structural biology is here to stay
As well as creating the tool itself, DeepMind has made policy decisions that have played a significant part in the transformation in structural biology. This includes its decision last July to make the code underlying AlphaFold open source, so that anyone can use the tool. Earlier this year, the company went further and lifted a restriction that hampered some commercial uses of the program.
It has also helped to establish, and is financially supporting, the AlphaFold database maintained with EMBL-EBI. DeepMind chief executive Demis Hassabis, his team, and their external collaborators deserve to be commended for this commitment to open science.
Last month, the company announced that it is establishing a research lab at the Francis Crick Institute, a flagship biomedical research centre in London. This is another welcome move, which will help to create and strengthen the close partnerships that are needed between researchers specializing in computational methods and those working more with hands-on tools.
AlphaFold on its own has limitations, as its designers fully acknowledge. For example, it is not designed to predict how a protein’s shape is altered by disease-causing mutations. It was also not originally intended to predict how proteins change shape when they interact with other proteins — although researchers are making progress on this next-generation challenge. And it’s not yet clear whether AlphaFold’s predictions will reliably provide the fine-grained detail necessary for drug discovery, such as the precise shape of the area on a protein to which a small molecule might bind — the kind of information that researchers in drug development crave.
Hassabis said last week that AlphaFold’s arrival will “require quite a big change in thinking”. That is starting to happen among researchers who are finding ways to use the tool, and are building on its insights.
But this change in thinking must also involve more companies and researchers, too, committing to open data, and to open-source software. Tomorrow’s applications, just like today’s AI tools, will not happen without terabytes of publicly accessible research, in various repositories, that software can learn from.

 ‘The entire protein universe’: AI predicts shape of nearly every known protein
‘The entire protein universe’: AI predicts shape of nearly every known protein
 What's next for AlphaFold and the AI protein-folding revolution
What's next for AlphaFold and the AI protein-folding revolution
 Protein-structure prediction revolutionized
Protein-structure prediction revolutionized
 Artificial intelligence in structural biology is here to stay
Artificial intelligence in structural biology is here to stay
 Read the paper: Highly accurate protein structure prediction for the human proteome
Read the paper: Highly accurate protein structure prediction for the human proteome
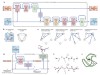 Read the paper: Highly accurate protein structure prediction with AlphaFold
Read the paper: Highly accurate protein structure prediction with AlphaFold