- NEWS AND VIEWS
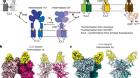
Receptor–enzyme complex structures show how receptors start to switch off
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 595, 499-500 (2021)
doi: https://doi.org/10.1038/d41586-021-01873-4
References
Chen, Q. et al. Nature 595, 600–605 (2021).
Nobles, K. N. et al. Sci. Signal. 4, ra51 (2011).
He, Y. et al. Cell Res. 27, 728–747 (2017).
Komolov, K. E. et al. Cell 169, 407–421 (2017).
Weis, W. I. & Kobilka, B. K. Annu. Rev. Biochem. 87, 897–919 (2018).
Gao, Y. et al. Mol. Cell 75, 781–790 (2019).
Kang, Y. et al. Nature 523, 561–567 (2015).
Cato, M. C. et al. Biomolecules 11, 447 (2021).
Palczewski, K., Buczylko, J., Kaplan, M. W., Polans, A. S. & Crabb, J. W. J. Biol. Chem. 266, 12949–12955 (1991).
Competing Interests
The authors declare no competing interests.

 Read the paper: Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1
Read the paper: Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1
 A self-activating orphan receptor
A self-activating orphan receptor
 Isoforms of GPCR proteins combine for diverse signalling
Isoforms of GPCR proteins combine for diverse signalling