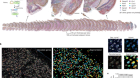
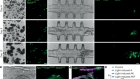
- TECHNOLOGY FEATURE
When antibodies mislead: the quest for validation
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 585, 313-314 (2020)
doi: https://doi.org/10.1038/d41586-020-02549-1
References
Laflamme, C. et al. eLife 8, e48363 (2019).
Andersson, S. et al. Nature Commun. 8, 15840 (2017).
Voskuil, J. L. A. et al. mAbs 12, 1794421 (2020).
Uhlen, M. et al. Nature Meth. 13, 823–827 (2016).
Menke, J., Roelandse, M., Ozyurt, B., Martone, M. & Bandrowski, A. Preprint at bioRxiv https://doi.org/10.1101/2020.01.15.908111 (2020).

 Antibody anarchy: a call to order
Antibody anarchy: a call to order
 Change-makers bring on recombinant antibodies
Change-makers bring on recombinant antibodies
 Reproducibility crisis: Blame it on the antibodies
Reproducibility crisis: Blame it on the antibodies
 NatureTech hub
NatureTech hub