- NEWS AND VIEWS
A decade of questions about the fluidity of cell identity
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 578, 522-524 (2020)
doi: https://doi.org/10.1038/d41586-020-00479-6
References
Vierbuchen, T. et al. Nature 463, 1035–1041 (2010).
Mangold, O. & Spemann, H. Wilhelm Roux Arch. Entwickl. Mech. Org. 111, 341–422 (1927).
Gurdon, J. B. J. Embryol. Exp. Morphol. 10, 622–640 (1962).
Davis, R. L., Weintraub, H. & Lassar, A. B. Cell 51, 987–1000 (1987).
Heins, N. et al. Nature Neurosci. 5, 308–315 (2002).
Buffo, A. et al. Proc. Natl Acad. Sci. USA 102, 18183–18188 (2005).
Barker, R. A., Götz, M. & Parmar, M. Nature 557, 329–334 (2018).
Xu, J., Du, Y. & Deng, H. Cell Stem Cell 16, 119–134 (2015).
Takahashi, K. & Yamanaka, S. Cell 126, 663–676 (2006).
Pang, Z. P. et al. Nature 476, 220–223 (2011).
Caiazzo, M. et al. Nature 476, 224–227 (2011).
Son, E. Y. et al. Cell Stem Cell 9, 205–218 (2011).
Victor, M. B. et al. Neuron 84, 311–323 (2014).
Mertens, J. et al. Cell Stem Cell 17, 705–718 (2015).
Hu, W. et al. Cell Stem Cell 17, 204–212 (2015).
Victor, M. B. et al. Nature Neurosci. 21, 341–352 (2018).
Wapinski, O. L. et al. Cell 155, 621–635 (2013).
Rouaux, C. & Arlotta, P. Nature Cell Biol. 15, 214–221 (2013).
Gascon, S., Masserdotti, G., Russo, G. L. & Götz, M. Cell Stem Cell 21, 18–34 (2017).
von Schimmelmann, M. et al. Nature Neurosci. 19, 1321–1330 (2016).

 Read the paper: Direct conversion of fibroblasts to functional neurons by defined factors
Read the paper: Direct conversion of fibroblasts to functional neurons by defined factors
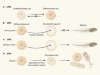 Cell identity reprogrammed
Cell identity reprogrammed
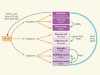 Cell reprogramming gets direct
Cell reprogramming gets direct