Reinventing the wheel is pointless, but for computational biologists it’s sometimes unavoidable. So when Rob Finn and Folker Meyer realized how much their work overlapped, they decided to try something different.
Finn is head of the sequence-families team at the European Bioinformatics Institute (EBI) in Hinxton, UK; Meyer is a computer scientist at Argonne National Laboratory in Lemont, Illinois. Both run facilities that let researchers perform a computationally intensive process called metagenomic analysis, which allows microbial communities to be reconstructed from shards of DNA. It would be helpful, they realized, if they could try each other’s code. The problem was that their analytical ‘pipelines’ — the carefully choreographed computational steps required to turn raw data into scientific knowledge — were written in different languages. Meyer’s team was using an in-house system called AWE, whereas Finn was working with nearly 9,500 lines of Python code.
“It was a horrible Python code base,” says Finn — complicated, and difficult to maintain. “Bits had been bolted on in an ad hoc fashion over seven years by at least four different developers.” And it was “heavily tied to the compute infrastructure”, he says, meaning it was written for specific computational resources and a particular way of organizing files, and thus essentially unusable outside the EBI. Because the EBI wasn’t using AWE, the reverse was also true. Then Finn and Meyer learnt about the Common Workflow Language (CWL).
CWL is a way of describing analytical pipelines and computational tools — one of more than 250 systems now available, including such popular options as Snakemake, Nextflow and Galaxy. Although they speak different languages and support different features, these systems have a common aim: to make computational methods reproducible, portable, maintainable and shareable. CWL is essentially an exchange language that researchers can use to share pipelines for whichever system. For Finn, that language brought sanity to his codebase, reducing it by around 73%. Importantly, it has made it easier to test, execute and share new methods, and to run them on the cloud.
There is a learning curve to adopting workflow languages. But, says Brian Naughton, data lead and co-founder of the drug-discovery firm Hexagon Bio in Menlo Park, California, “the energy that you expend learning is more than made up for by the energy you save in having your code be reproducible.”
Step by step
For computational biologists, pipelines are methods; much like wet-lab protocols, they must be documented. But pipelines often comprise dozens of steps, so it’s not trivial to do. Bioinformatician Titus Brown at the University of California, Davis, calculated that passing six samples through his de novo transcriptome assembly pipeline — involving data download, quality control, normalization, assembly, annotation and analysis — requires “well over 100 steps”. Researchers must document precisely how each step is performed if they have any hope of reproducing them at a later date.
Typically, researchers codify workflows using general scripting languages such as Python or Bash. But these often lack the necessary flexibility. Workflows can involve hundreds to thousands of data files; a pipeline must be able to monitor their progress and exit gracefully if any step fails. And pipelines must be smart enough to work out which tasks need to be re-executed and which do not.
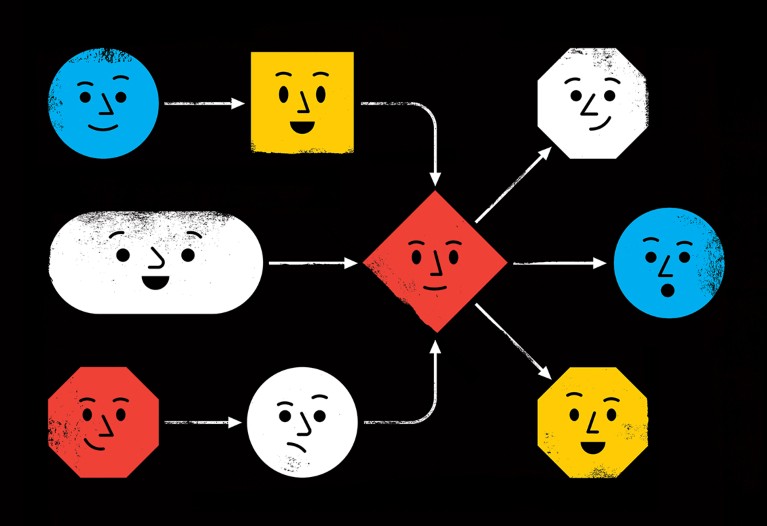
Bioinformatician Davis McCarthy at St Vincent’s Institute of Medical Research in Fitzroy, Australia, says Python and R were more than enough for the relatively simple workflows he used as a PhD student. But today, McCarthy, who works with single-cell data sets, processes orders of magnitude more samples, some of which inevitably fail owing to problems such as network issues and memory shortages. “It was just way beyond my capabilities to figure that out from scratch for an analysis of this size,” he says. He adopted the command-line-driven Snakemake, instead (see ‘Anatomy of a workflow’).
More problematic is scalability: a script that works on a laptop can rarely run on a computing cluster or the cloud without modification. These systems typically have specific formatting, authentication and configuration requirements to describe, for instance, the necessary computing resources. And then there’s the fact that many pipelines are tightly tailored to a lab’s specific computational environment.
Workflow systems can ease that complexity. Some, such as Galaxy, allow users to build pipelines in a point-and-click user interface; others operate at the text-based command line. Yet all typically support a few core functions, including ‘re-entrancy’ (picking up where the workflow left off in an earlier run), scalability and the ability to specify the computational environment for each step. Many support software-installation tools such as Conda, as well as containerization systems such as Docker, which means that other users can run those pipelines, even if their own computing infrastructure differs. And workflow systems can generate detailed reports of what they did.
Containers are particularly powerful additions to workflows, says Carole Goble, a computer scientist at the University of Manchester, UK, because they allow researchers to package the precise computational components required to execute each step; other users can then download those containers to recreate that environment on their own system. They even allow for different steps to use components that are not mutually compatible, such as different versions of the Python programming language. “Thank God for containers, because that’s made an enormous difference,” she says.
In 2018, researchers in the African bioinformatics network H3ABioNet built four pipelines — two each in CWL and Nextflow — and embedded them in Docker containers (S. Baichoo et al. BMC Bioinform. 19, 457; 2018). Doing so made the results reproducible and the workflows accessible across the network, despite differing computational resources.
Gold standard
Ruchi Munshi, a software-product manager at the Broad Institute of MIT and Harvard in Cambridge, Massachusetts, says computational biologists there were motivated to develop and adopt the Cromwell workflow system and its language WDL (Workflow Definition Language) to provide better standardization and sharing of workflows across the institute, greater accessibility for non-programmer biologists and scalability.
Standardization, Munshi notes, is a particular benefit of workflow systems. With scripting languages, there is no single way to write a pipeline, making it difficult to cobble together different steps into a new pipeline. But using a workflow language, researchers can create libraries of compatible pieces that can be assembled like building blocks. The Broad Institute, for instance, publishes WDL-based pipelines for its GATK software on GitHub and Terra, and at the containers registry dockstore.org. (Dockstore also lists workflows written in Nextflow and CWL.)
The ‘nf-core’ project is creating a set of gold-standard bioinformatics pipelines for the Nextflow system. Philip Ewels, who founded nf-core at SciLifeLab in Stockholm, says its goal is to create pipelines that work in a consistent way and support a uniform set of features, such as Docker and Conda compatibility. “If you can use one of these pipelines, you can use all of them,” he says.
CWL, by contrast, aims for portability across workflow systems, by separating the computational parts of a workflow from the code required to execute them. Goble, who is building a searchable registry of CWL workflows, says: “The CWL’s job is really to be the workflow language in common. And that’s it. It does exactly what it says on the tin.”
So, do you need a workflow system? Not every task requires one, and there is a learning curve. Scripting usually suffices for one-off tasks and when working out the pipeline itself. The tipping point, most agree, comes when you need to run the same workflow over and over again, or if the data are likely to be published.
Fortunately, says Ewels, the languages are easy to pick up, and examples are abundant. “Once you’re familiar with it, you get in the habit of using it,” he says. “And then you wonder how you ever lived without it.”


 Reproducibility of computational workflows is automated using continuous analysis
Reproducibility of computational workflows is automated using continuous analysis
 Software simplified
Software simplified
 Cloud computing for genomic data analysis and collaboration
Cloud computing for genomic data analysis and collaboration
 NatureTech hub
NatureTech hub