- NEWS AND VIEWS
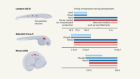
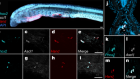
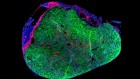
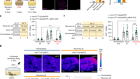
What makes flatworms go to pieces
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 572, 593-594 (2019)
doi: https://doi.org/10.1038/d41586-019-02376-z
References
Arnold, C. P., Benham-Pyle, B. W., Lange, J. J., Wood, C. J. & Sánchez Alvarado, A. Nature 572, 655–659 (2019).
Steinhart, Z. & Angers, S. Development 145, dev146589 (2018).
Wiese, K. E., Nusse, R. & van Amerongen, R. Development 145, dev165902 (2018).
Gurley, K. A., Rink, J. C. & Sánchez Alvarado, A. Science 319, 323–327 (2008).
Niehrs, C. Development 137, 845–857 (2010).
Stuckemann, T. et al. Dev. Cell 40, 248–263 (2017).
Best, J. B., Goodman, A. B. & Pigon, A. Science 164, 565–566 (1969).
Malinowski, P. T. et al. Proc. Natl Acad. Sci. USA 114, 10888–10893 (2017).
Petersen, C. P. & Reddien, P. W. Cell 139, 1056–1068 (2009).
Molina, M. D., Salo, E. & Cebria, F. Dev. Biol. 311, 79–94 (2007).
Collins, J. J. III et al. PLoS Biol. 8, e1000509 (2010).
Cowles, M. W., Omuro, K. C., Stanley, B. N., Quintanilla, C. G. & Zayas, R. M. PLoS Genet. 10, e1004746 (2014).
Kobayashi, C., Saito, Y., Ogawa, K. & Agata, K. Dev. Biol. 306, 714–724 (2007).
Reddien, P. W. Cell 175, 327–345 (2018).
Sikes, J. M. & Bely, A. E. Dev. Biol. 338, 86–97 (2010).
Cannon, J. T. et al. Nature 530, 89–93 (2016).
Zattara, E. E. & Bely, A. E. Evol. Dev. 13, 80–95 (2011).
Burton, P. M. & Finnerty, J. R. Dev. Genes Evol. 219, 79–87 (2009).
Chapman, J. A. et al. Nature 464, 592–596 (2010).
Petersen, H. O. et al. Mol. Biol. Evol. 32, 1928–1947 (2015).

 Read the paper: Wnt and TGFβ coordinate growth and patterning to regulate size-dependent behaviour
Read the paper: Wnt and TGFβ coordinate growth and patterning to regulate size-dependent behaviour
 On with their heads
On with their heads
 Regeneration writ large
Regeneration writ large