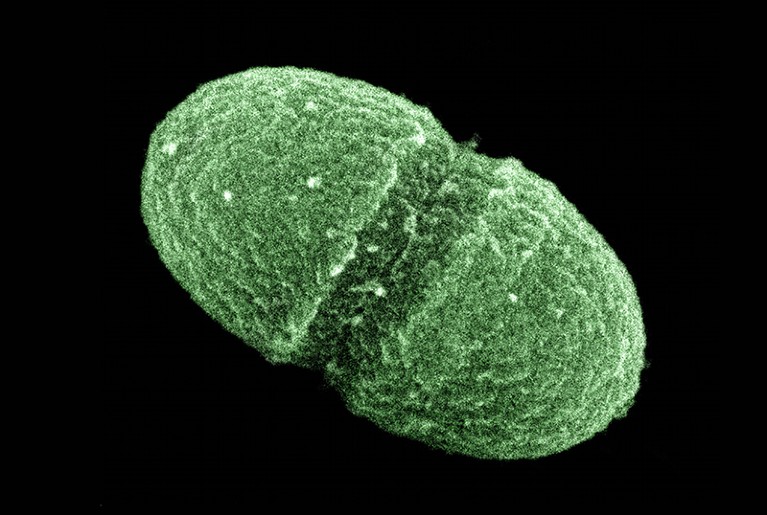
The gut bacterium Enterococcus faecalis is one of many thousands of microorganisms under investigation by the ongoing Human Microbiome Project.Credit: Media for Medical/UIG/Getty
Twenty years ago, the human microbiome — or the community of microorganisms living in the body — was a fledgling field. Now, it is a flourishing area of research that integrates the basic and clinical sciences, and continues to attract large sums of public and private investment across the globe. One of the first large-scale initiatives was the US National Institutes of Health (NIH)-funded 10-year Human Microbiome Project (HMP), launched in 2007. One of its biggest initial revelations was that the taxonomic composition of the microbiota in the human body was not a reliable predictor of host phenotype, such as disease susceptibility. This was the impetus for a more comprehensive analysis of both the microbiome and the host, culminating in the second phase, the integrative HMP (iHMP). The key results of this project are published this week in Nature and Nature Medicine.
Nature Collection: Human Microbiome Project, part 2
One of the Nature studies examines the gut microbiome of people with inflammatory bowel disease and finds that community composition and immune responses are significantly less stable in individuals with this disease. In another study, the authors analysed human gut and nasal microbiomes during the onset of type 2 diabetes and show that microbial and host profiling can together predict insulin sensitivity status, despite high microbiome variability between individuals. The third study found that in pregnant women, vaginal microbiome profiles before 24 weeks’ gestation provide a marker for risk of premature birth, particularly among women of African ancestry (J. M. Fettweis et al. Nature Med. https://doi.org/10.1038/s41591-019-0450-2; 2019).
A major strength of all three studies is their unprecedented depth and breadth of molecular data for both the host and the microbiota. Another is their longitudinal design, which provides important insights into how host–microbiota interactions change over time.
So, what lies ahead now that the second phase of this major project has come to an end? Many questions about the basic biology of the microbiota remain, including what drives its variation over time and between populations and geographic regions. Ultimately, the goal is to translate such findings into clinical interventions — a monumental challenge. This will require close multidisciplinary collaboration. For example, the microbiology community on its own is unlikely to identify the animal models that are most appropriate for investigating a particular medical condition, or to establish the minimum criteria for substantiating claims of causality.
Multidisciplinary efforts require time and sustained funding to foster innovative ideas and drive translational research. A field this big and mature would benefit from a central agency or a dedicated institute to foster the necessary multidisciplinary collaborations and to focus on standardization, including data sharing and best practices, as well as on the ethical, regulatory and societal implications of such studies. As Lita Proctor, former HMP coordinator at the US National Human Genome Research Institute, discusses, there are lessons to be learnt from other disciplines such as ocean sciences.
To build on the achievements of the microbiome community so far, strong leadership and coordination are a priority. There are encouraging signs that the field is moving in this direction.

 Nature Collection: Human Microbiome Project, part 2
Nature Collection: Human Microbiome Project, part 2
 Read the paper: The Integrative Human Microbiome Project
Read the paper: The Integrative Human Microbiome Project
 Priorities for the next 10 years of human microbiome research
Priorities for the next 10 years of human microbiome research
 News & Views: Tracking humans and microbes
News & Views: Tracking humans and microbes
 Read the paper: Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases
Read the paper: Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases
 Read the paper: Longitudinal multi-omics of host–microbe dynamics in prediabetes
Read the paper: Longitudinal multi-omics of host–microbe dynamics in prediabetes
 Read the paper: The vaginal microbiome and preterm birth
Read the paper: The vaginal microbiome and preterm birth
 Read the paper: Racioethnic diversity in the dynamics of the vaginal microbiome during pregnancy
Read the paper: Racioethnic diversity in the dynamics of the vaginal microbiome during pregnancy