Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 569, 340-342 (2019)
doi: https://doi.org/10.1038/d41586-019-01407-z
Updates & Corrections
-
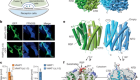
Correction 13 May 2019: An earlier version of this article said that only 5 of the 11 thiol groups of an undecameric protein participate in metal-mediated connections to assemble into a pentagonal icositetrahedron. The correct number is 10 of the 11. The main text, figure and caption have been corrected accordingly.
References
Yeates, T. O. Annu. Rev. Biophys. 46, 23–42 (2017)
Aumiller, W. M., Uchida, M. & Douglas, T. Chem. Soc. Rev. 47, 3433–3469 (2018).
Malay, A. D. et al. Nature 569, 439–443 (2019).
Holden, A. Shapes, Space, and Symmetry (Columbia Univ. Press, 1971).
Padilla, J. E., Colovos, C. & Yeates, T. O. Proc. Natl Acad. Sci. USA 98, 2217–2221 (2001).
King, N. P. et al. Science 336, 1171–1174 (2012).
Lai, Y.-T. et al. Nature Chem. 6, 1065–1071 (2014).
Sciore, A. et al. Proc. Natl Acad. Sci. USA 113, 8681–8686 (2016).
Bale, J. B. et al. Science 353, 389–394 (2016).
Basta, T. et al. Proc. Natl Acad. Sci. USA 111, 670–674 (2014).
Salgado, E. N., Radford, R. J. & Tezcan, F. A. Acc. Chem. Res. 43, 661–672 (2010).
Caspar, D. L. & Klug, A. Cold Spring Harb. Symp. Quant. Biol. 27, 1–24 (1962).
Sasaki, E. et al. Nature Commun. 8, 14663 (2017).

 Read the paper: An ultra-stable gold-coordinated protein cage displaying reversible assembly
Read the paper: An ultra-stable gold-coordinated protein cage displaying reversible assembly
 Machine learning speeds up synthesis of porous materials
Machine learning speeds up synthesis of porous materials
 Designer protein delivers signal of choice
Designer protein delivers signal of choice