- NEWS AND VIEWS
Elusive microbe that consumes ethane found under the sea
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 568, 40-41 (2019)
doi: https://doi.org/10.1038/d41586-019-00842-2
References
Judd, A. G. Environ. Geol. 46, 988–996 (2004).
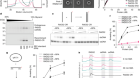
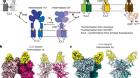
Chen, S.-C. et al. Nature 568, 108–111 (2019).
Krukenberg, V. et al. Environ. Microbiol. 20, 1651–1666 (2018).
Laso-Pérez, R. et al. Nature 539, 396–401 (2016).
McGlynn, S. E., Chadwick, G. L., Kempes, C. P. & Orphan, V. J. Nature 526, 531–535 (2015).
Milucka, J. et al. Nature 491, 541–546 (2012).
Ermler, U., Grabarse, W., Shima, S., Goubeaud, M. & Thauer, R. K. Science 278, 1457–1462 (1997).
Wongnate, T. et al. Science 352, 953–958 (2016).
Ragsdale, S. W. & Pierce, E. Biochim. Biophys. Acta 1784, 1873–1898 (2008).
Plass-Dülmer, C., Koppmann, R., Ratte, M. & Rudolph, J. Glob. Biogeochem. Cycles 9, 79–100 (1995).
Etiope, G. & Ciccioli, P. Science 323, 478 (2009).
Bousquet, P. et al. Nature 443, 439–443 (2006).
Dalsøren, S. B. et al. Nature Geosci. 11, 178–184 (2018).

 Read the paper: Anaerobic oxidation of ethane by archaea from a marine hydrocarbon seep
Read the paper: Anaerobic oxidation of ethane by archaea from a marine hydrocarbon seep
 Forty years of fathoming life in hot springs on the ocean floor
Forty years of fathoming life in hot springs on the ocean floor
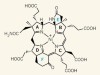 Origin of a key player in methane biosynthesis
Origin of a key player in methane biosynthesis