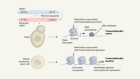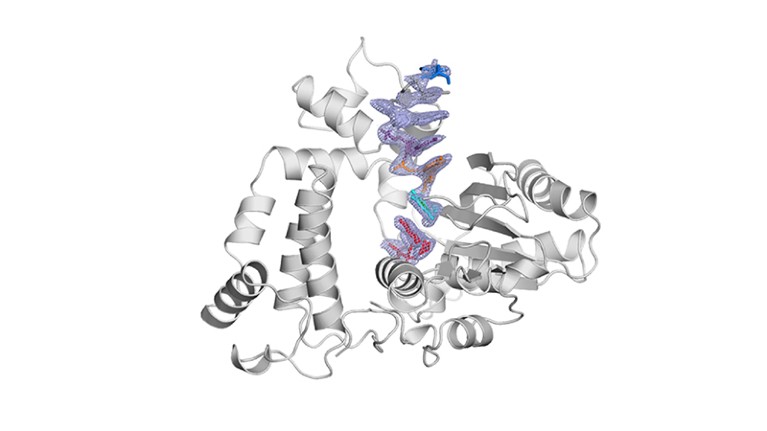
TdT, a template-independent DNA polymerase.Credit: Marc Delarue, adapted from J. Gouge et al. J. Mol. Biol. 425, 4334–4352 (2013).
For decades, biologists have built custom DNA sequences chemically, from phosphoramidite building blocks that replicate natural bases. But the method is impractical beyond 200 bases, and environmentally hazardous. New enzymatic strategies could circumvent those limitations.
In June 2018, George Church, a geneticist based at Harvard University in Cambridge, Massachusetts, and his colleagues reported encoding and decoding short messages in enzymatically synthesized DNA (H. H. Lee et al. Preprint at bioRxiv https://doi.org/c2cs; 2018); two months later, Molecular Assemblies, a biotechnology company in San Diego, California, announced a similar achievement.
In July, Sebastian Palluk and Daniel Arlow, in Jay Keasling’s synthetic-biology laboratory at the University of California, Berkeley, published a strategy that they used to build ten-base oligonucleotides (S. Paluk et al. Nature Biotechnol. http://doi.org/gdqkff; 2018), and founded Ansa Biotechnologies to commercialize the approach.
And in October, DNA Script, based in Paris, announced that it had synthesized a 150-base DNA strand of defined sequence — an achievement that William Efcavitch, Molecular Assemblies’ chief scientific officer, calls a “milestone”. (At least two other companies also are pursuing enzymatic strategies: Nuclera Nucleics and Evonetix, both based near Cambridge, UK.)
Key to enzymatic synthesis is terminal deoxynucleotidyl transferase (TdT), a DNA polymerase that requires no template. “It can add nucleotides without taking instructions,” explains Marc Delarue, a structural biologist at the Pasteur Institute in Paris who collaborates with DNA Script. In theory, the approach can generate longer molecules than can chemical synthesis. It’s also environmentally friendlier.
To control the sequence, developers must stop the enzyme after each step. Ansa tethers the nucleotide to the enzyme, thus physically blocking the DNA; others are developing TdT variants and modified DNA bases that act as reversible terminators. For DNA-based information storage, in which data are encoded in the transitions between bases rather than in their precise arrangement, the native enzyme and nucleotides can be used.
Enzyme-written DNAs are not yet commercially available. Nor can any published strategy rival chemical synthesis in length or efficiency. Palluk and Arlow reported 97.7% average coupling efficiency in their paper; Integrated DNA Technologies (IDT), a DNA-synthesis firm in Coralville, Iowa, touts 99.5%. Yet less than 40% of molecules are correct at 200 bases; longer molecules would require higher efficiencies.
Still, says Emily Leproust, chief executive of the synthetic-DNA firm Twist Bioscience in San Francisco, California, “someone will crack it, and it’s going to be great for the field”. Adam Clore, technical director of synthetic biology at IDT, reckons that a “commercially viable product” is “probably several years off”.
Those products could fill niches that chemistry cannot: long, complex sequences — synthetic gene libraries, for instance — for which assembly from shorter segments can add significant delays. “Any technology that can make that faster is going to be very valuable,” says Christopher Voigt, a synthetic biologist at the Massachusetts Institute of Technology in Cambridge. “There is no Nobel prize that needs to happen,” Leproust says. “It’s just hard engineering.”

 The automatic-design tools that are changing synthetic biology
The automatic-design tools that are changing synthetic biology
 How to hack the genome
How to hack the genome