- NEWS AND VIEWS
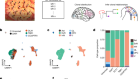
Chromosomes come together to help mice distinguish odours
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 565, 439-440 (2019)
doi: https://doi.org/10.1038/d41586-019-00010-6
References
Bushdid, C., Magnasco, M. O., Vosshall, L. B. & Keller, A. Science 343, 1370–1372 (2014).
Rodriguez, I. Cell 155, 274–277 (2013).
Monahan, K., Horta, A. & Lomvardas, S. Nature 565, 448–453 (2019).
de Laat, W. & Duboule, D. Nature 502, 499–506 (2013).
Dixon, J. R. et al. Nature 485, 376–380 (2012).
Spitz, F. Semin. Cell Dev. Biol. 57, 57–67 (2016).
Schoenfelder, S. et al. Nature Genet. 42, 53–61 (2010).
Zhang, Y. et al. Nature 504, 306–310 (2013).
Lomvardas, S. et al. Cell 126, 403–413 (2006).
Fuss, S. H., Omura, M. & Mombaerts, P. Cell 130, 373–384 (2007).
Markenscoff-Papadimitriou, E. et al. Cell 159, 543–557 (2014).
Monahan, K. et al. eLife 6, e28620 (2017).
Armelin-Correa, L. M., Gutiyama, L. M., Brandt, D. Y. C. & Malnic, B. Proc. Natl Acad. Sci. USA 111, 2782–2787 (2014).
Crocker, J. et al. Cell 160, 191–203 (2015).
Noordermeer, D. et al. Nature Cell Biol. 13, 944–951 (2011).
Johnston, R. J. & Desplan, C. Science 343, 661–665 (2014).

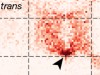 Read the paper: LHX2- and LDB1-mediated trans interactions regulate olfactory receptor choice
Read the paper: LHX2- and LDB1-mediated trans interactions regulate olfactory receptor choice
 Neural circuit evolved to process pheromone differently in two species of fruit fly
Neural circuit evolved to process pheromone differently in two species of fruit fly
 A mixed model of neuronal diversity
A mixed model of neuronal diversity