When Yvonne Chen published the first paper1 on a particular immune cell engineered to target either of two protein fragments on a cancer cell, several colleagues tried to discourage her from describing her creation in the unfamiliar language of computer logic. She did it anyway.
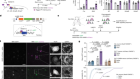
Chen, a chemical and biomolecular engineer at the University of California, Los Angeles, had used synthetic proteins known as chimaeric antigen receptors (CARs) to modify the immune cell, a T lymphocyte, so that it could look for the two fragments — CD19 and CD20, examples of immune-system-stimulating molecules called antigens. This design meant that, if the cancer it was attacking underwent a mutation that rendered one antigen unrecognizable, the T cell could still use the other one to find and kill the cancer cell. Instead of referring to her creation using the existing biological terminology, as a bispecific cell, she called it an OR-gate CAR T cell, because of its ability to recognize one target or the other. “I actually received advice from multiple people saying, ‘You shouldn’t call it an OR-gate CAR, because people who work on T cells, who are either cell biologists or physicians, won’t understand what an OR gate means’,” she says. “But now everybody calls it an OR-gate CAR, because it makes sense.”
The concept of an OR gate is more familiar in computer science, where it refers to a logical operation that is triggered in the presence of either of two input options. But it describes in a few letters what the cell actually does, and distinguishes it from other types of bispecific CAR T cell, Chen says. Her OR-gate cell is nearly ready to begin clinical trials, and her lab is developing cells that mimic other logical functions, such as AND, which operates only if two inputs are positive, and NOT, which gives a negative output if the input is positive.
Merging fields
Terminology and concepts from computer science and engineering are becoming more common in biology labs as scientists re-engineer the activity of cells for specific applications. They are gaining unprecedented capabilities both from genetic-editing tools that have been around for a while, such as those that use viruses or proteins called zinc fingers, and from new CRISPR–Cas9 technology that allows more-targeted editing of DNA.
Scientists are creating vast collections of data by tweaking the transcription factors that copy DNA, one at a time and in various combinations, to see how each changes the cell. This produces so many variations and so much complexity that it calls for a computer scientist, who can build a model of what’s going on. Other researchers are creating potential therapies, some based on a patient’s own immune cells, and allowing a fresh understanding of embryonic development. And they might one day enable people to create products now undreamt of as humans gain, in the words of Drew Endy, a bioengineer at Stanford University in California, “mastery of living matter”.
Illustration by David Parkins
Chen developed her OR-gate CAR to tackle tumour escape, in which a cancer mutates and becomes unrecognizable to the immune therapy attacking it. With her T cell, the cancer would have to lose two antigens to become invisible to the immune system — a much less likely event. But CAR T-cell therapy can have the opposite problem: when targeting certain types of cancer, it can recognize a similar antigen on a healthy cell and so attack that, too. So Chen has also devised a different type of T cell as a biological AND gate. In that system, when the T cell receives a signal from the target antigen, it expresses a second receptor. Only if that second receptor also finds its own antigen on the target cell does the T cell activate and attack. Which type of T cell to use, AND or OR, would be determined by the characteristics of the cancer being treated.
There’s even more that might be done using cellular logic, says Endy. “Today, we have the full set of Boolean logic operators, operating in a diversity of cell types, implemented with a diversity of molecular mechanisms, and it’s just getting better and better,” he says. He imagines programming a cell to count its own divisions. If some cells start dividing too rapidly, that might be an early sign of cancer, and triggering programmed cell death could nip a tumour in the bud before it’s even big enough to be detected by other means.
Another Stanford researcher, pathologist Marius Wernig at the Institute for Stem Cell Biology and Regenerative Medicine, envisions the creation of ‘smart cells’ that could monitor the body for all sorts of disease processes, and take action if something goes awry. Such a creation is a long way off, he thinks, but not impossible. “We are at a really exciting time,” says Wernig, because CRISPR and other genetic engineering tools “really open up tremendous possibilities”.
Wernig’s main focus is on regenerative medicine. His lab was the first to turn cells that normally generate skin tissue into functional neurons2. In particular, he is using stem cells generated from adult cells to develop a treatment for dystrophic epidermolysis bullosa, a genetic disease that causes the skin to blister and crack. He aims to harvest cells from patients, convert them into stem cells, modify them genetically and then turn them back into skin that he can use as a graft to replace damaged tissue. He hopes that the treatment will enter clinical trials within two or three years.
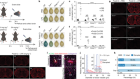
To get an understanding of how editing the mechanisms in a cell changes the cell’s behaviour, Wernig and his colleagues used CRISPR to change factors individually, then in combination. Instead of clipping out or adding a bit of DNA to a cell’s genome, he turned transcription factors in a human cell on or off to see what effect that had. He did that for more than 2,000 transcription factors, as well as some DNA-tweaking enzymes called chromatin modifiers, essentially pushing every lever in the cell’s machinery one at a time to see what happened3.
That systematic engineering approach is a new way of finding answers in biology, says Patrick Cahan, a computational biologist in the Institute of Cell Engineering at Johns Hopkins University in Baltimore, Maryland. “They can watch large combinations of genes turn on and turn off, irrespective of what we thought we knew about those genes from decades of developmental biology,” Cahan says. “It’s just, ‘Let’s see what’s possible’.”
Processing power
Computer science has a big role in cellular engineering, Cahan says, in part because experiments such as Wernig’s generate enormous amounts of data. When biologists perform an assay to look at which genes are expressed in particular cells, which are not and in what abundance, the result can be a data set containing typically 20,000–30,000 variables across thousands of individual cells. Making sense of it all, especially when many different factors are working together in complex combinations, requires computer modelling and machine learning.
Cahan aims to make sure that all of his students develop both a sense of comfort with and a sense of scepticism towards genome-scale computing. The comfort comes in feeling assured that computing can provide valuable answers. The scepticism has to do with recognizing which questions the data cannot answer, and perhaps designing a study from the outset to make sure that the researchers are getting data that will allow them to find the answers they are seeking.
It can be too easy for inexperienced researchers to organize the data to fit their hypothesis, and then fool themselves into thinking they are seeing something they are not. But they can also make the opposite mistake. “When you see something that is inconsistent with your hypothesis, you might think it’s some sort of artefact of this large-scale data set and you might ignore it,” Cahan says, “and that might be the gem.”
Although people have become very good at manufacturing inanimate objects ranging from computer chips to cars, and even at making relatively simple biological products, such as drugs called monoclonal antibodies, the industrial production of cells is a new area entirely, says Krishnendu Roy, a biomedical engineer who directs the National Science Foundation’s Engineering Research Center for Cell Manufacturing Technologies at the Georgia Institute of Technology in Atlanta. “For the first time probably in human history, we are trying to do industrial-scale manufacturing of a living product,” Roy says. “The whole paradigm of manufacturing needs to change.”
One major challenge is that living cells tend to change depending on their environment. Different batches of reagents, container materials, whether they are in a 2D or 3D structure and even the presence of electrical fields can alter which genes are triggered, which proteins are expressed and which metabolites are produced.
“We have a very sensitive product that changes with slight manipulation. Whether those changes are important or not important is something we still need to figure out,” Roy says. So engineers need to understand the biological processes, and biologists need to understand the industrial-production systems. “If you just put together a bunch of engineers and give them the cells, they’re not going to be able to solve this,” says Roy.
Expanded horizons
Cellular engineering is a multidisciplinary field, and it is important for researchers to be literate in all the specialities that touch on their work. “Those fields of cell biology and health care and data science are really merging now to give us insights of properties and functions that we really never had insights on,” Roy says.
Combining areas of expertise and ways of approaching problems from various fields — molecular biology, bioinformatics, chemical engineering, industrial engineering — is what makes cellular engineering function. Endy, who has helped to design undergraduate bioengineering courses at both Stanford and the Massachusetts Institute of Technology in Cambridge (see ‘Where science meets engineering’), says that scientists and engineers have their own way of looking at fundamental scientific questions. “For me as an engineer, it’s the making of this thing that works, whereas the end product of the biologist is knowledge, and a description of how biology works,” he says.
All the researchers say they had to learn a lot from outside their major field. Chen, for instance, had to learn how to run a clinical trial. She discovered that one concern is determining the volume of a drug that is practical to infuse into a patient, something she had never considered as a bench scientist. She also had to think about not just whether what she was trying to do worked, but whether she could do it in a way that was patentable — immune therapy is very expensive, so maintaining intellectual-property rights that can help to pay for the clinical trials is an important consideration.
In fact, Endy argues that the field opens up a whole host of not just scientific and engineering questions, but also questions of ethics and policy. “We have a capacity to enable 10 billion people to flourish without trashing the planet,” he says. “It looks to me we’re going to secure operational mastery of living matter, and we’re going to do that by about 2030, and we could do it faster. Then the question is, what do we wish of biology? What do we want of our relationship with biology? That is not a science and engineering question.”

 Nature special: Bottom-up biology
Nature special: Bottom-up biology
 Partner Content: A research home away from home
Partner Content: A research home away from home