THE PAPERS IN BRIEF
• Proteins called channelrhodopsins form light‑activated ion channels in cell membranes.
• Channelrhodopsins have been used for optogenetics — a revolutionary technique that uses light to induce ion flux through these channels in genetically engineered cells, and thereby controls physiological processes.
• So far, the channelrhodopsins used most widely for optogenetics conduct positively charged ions (cations).
• However, the use of channelrhodopsins that conduct negatively charged ions (anions) would open up new possibilities for optogenetics.
• Writing in Nature, Kim et al.1 and Kato et al.2 describe the crystal structures of two anion-conducting channelrhodopsins, revealing the molecular basis of light-gated anion conduction, and providing insight into how these proteins could be modified for optogenetics.
• Kato et al. also report an engineered anion-conducting channelrhodopsin suitable for use in optogenetics.
PATRICK SCHEERER: Structural insight
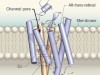
In the early 2000s, two channelrhodopsins were discovered3,4 in the microbial alga Chlamydomonas reinhardtii. Both are transmembrane receptors that have seven membrane-spanning α-helices and contain a deeply embedded molecule called retinal, which is covalently attached to the proteins, and is responsible for their light sensitivity. The proteins form channels that, when activated by light, allow various cations to flow down the electrochemical gradients that form across cell membranes when there are unequal concentrations of ions inside and outside the cell.
Read the paper: Crystal structure of the natural anion-conducting channelrhodopsin GtACR1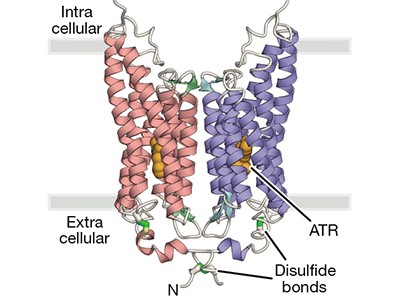
In 2012, the first crystal structure5 of a cation-conducting channelrhodopsin (CCR) was published — an engineered protein called C1C2, in which parts of the two C. reinhardtii channelrhodopsins were fused together. This breakthrough provided a snapshot of the structure of the light-activated channel and hinted at how cations are selectively conducted through it.
The C1C2 structure was subsequently used in conjunction with molecular modelling data to guide the design of engineered channelrhodopsins for use in optogenetics. As part of these efforts, several groups reported that certain structural modifications — including remodelling of the inner surface of the C1C2 pore, and specific mutations to the central part of the protein that acts as the ion gate — could make channelrhodopsins anion-selective. This resulted in the development of highly chloride-selective channelrhodopsins6–9, including a variant known as iC++, the structure of which is now reported by Kato and colleagues. A naturally occurring anion-conducting channelrhodopsin (GtACR1) was also discovered10 in the alga Guillardia theta, and its structure is reported by Kim and co-workers.
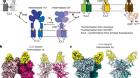
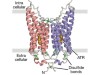
The structure of GtACR1 shows that this protein shares a similar overall architecture with CCRs, but has several key differences (Fig. 1). For example, most of the amino acids at the surface of the GtACR1 channel are positively charged (as is appropriate for cation exclusion), and not negatively charged as in CCRs. CCRs contain two extracellular vestibules (EV1 and EV2), only one of which (EV2) connects to the ion-conducting pathway. But in GtACR1 (and also in iC++), both vestibules are remodelled, and only EV1 is connected to the ion-conducting pathway.
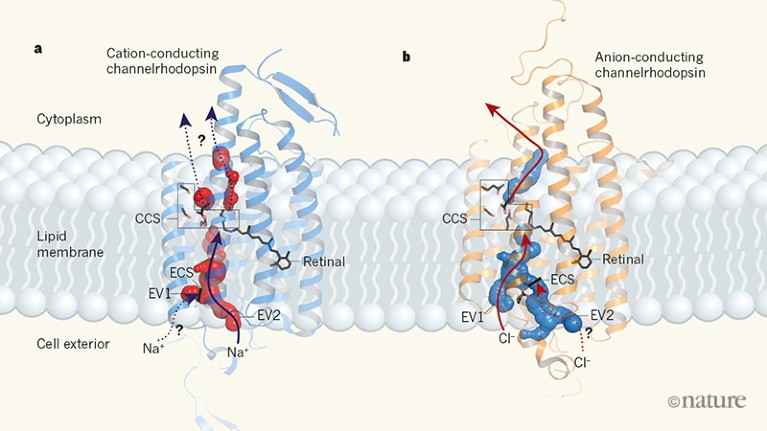
Figure 1 | The structures of cation-conducting and anion-conducting channelrhodopsin proteins. Channelrhodopsin proteins that transport positively charged ions (cations) such as sodium (Na+) into cells have been widely used as tools for optogenetics — a technique that allows neurons to be activated by light pulses, aided by a light-sensitive molecule called retinal. Two papers1,2 now report the structures of channelrhodopsins that conduct negatively charged ions (anions) such as chloride (Cl−) and the development of an engineered anion-conducting channelrhodopsin2 that could be used in optogenetics to inhibit neuronal activity. a, b, The structure5 of a cation-conducting channelrhodopsin called C1C2 (a) and of the anion-conducting channelrhodopsin GtACR1 reported by Kim et al.1 (b). Both proteins are shown in their closed conformation, in which the central ion-conducting channel is open, apart from a closed central constriction site (CCS). Some of the key amino-acid residues of the CCS are shown, represented in a stick format. Solid arrows trace a possible route that ions might take through this central channel to enter the cell, and the path they would take out of GtACR1 if the channel was open. Dotted arrows indicate other ion-transit pathways that might be used and that would be blocked by an extracellular constriction site (ECS) or the closed CCS. The channel-exit path between the CCS and the cytoplasm is clearly visible as a continuous path only in the anion-conducting channel. The central cation-conducting channel (dark red) in a is mostly negatively charged, whereas the central anion-conducting channel (dark blue) in b is mostly positively charged. Another structural difference between the channels is that the extracellular-vestibule (EV) region that connects to the central ion-conducting pathway is EV2 in the cation-conducting channel and EV1 in the anion-conducting channel.
Although the structure of GtACR1 captures the protein in its closed state, the channel is almost entirely open — remarkably, it is blocked by only one central constriction and at an extracellular constriction site in EV2. Kim et al. find that the central constriction contains three key amino-acid residues, two of which are involved in the process of anion transport, whereas the other is important for anion selectivity. A clear pathway allows anions released from the centre of the pore to reach the cell interior (although such release would not occur in this closed conformation). For comparison, in CCRs, the intracellular region of the protein is mainly obstructed owing to the closure of the channel.
Kim et al. and Kato et al. report further analyses of GtACR1 and iC++ that provide insight into other aspects of how the structure of anion-conducting channelrhodopsins affects their function. For example, in GtACR1, they find that residues along the central constriction, in the retinal-binding pocket and at an extracellular region of the protein greatly affect the kinetics of pore closing. The two papers, in combination with two other recently described crystal structures of natural channelrhodopsins11,12, provide a deeper structural and functional understanding of the light-activated ion-gating mechanism in microbial channelrhodopsins — and provide a basis for designing new classes of cation- and anion-conducting ion channels for optogenetics.
ELIZABETH UNGER & LIN TIAN: Tool development
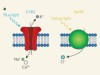
Neurons in the brain function primarily by generating complex patterns of electrical impulses called action potentials, which were, for a long time, extremely cumbersome to measure and even more difficult to manipulate. That changed in 2005, when channelrhodopsin was introduced as a tool for optogenetics13,14. When expressed in neurons, channelrhodopsin can be activated by light pulses to force those cells to fire action potentials. Optogenetics can precisely excite individual neurons or entire populations of genetically defined neurons in naturally behaving animals. This allowed direct testing of the contributions of different types of neuron to behavioural outcomes. The initial success of channelrhodopsin spawned an array of variants of the protein that have a range of useful properties for optogenetics15.
Read the paper: Structural mechanisms of selectivity and gating in anion channelrhodopsins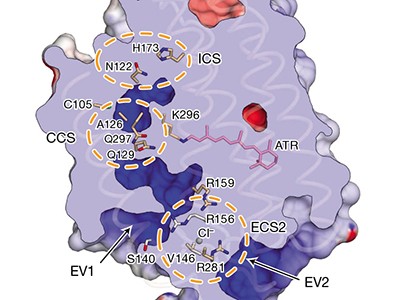
But just as much information is encoded by the absence of an action potential as by the presence of one. To fully understand the brain, we therefore need to be able to inhibit firing as well as to stimulate it — and to inhibit firing, proteins that move anions into cells in a manner that is controlled by light are needed. Unfortunately, optogenetic tools for inhibition have lagged considerably behind those for excitation. Light-activated proteins from the ion-pump family that could be used as inhibitory proteins were reported16 shortly after channelrhodopsin. But because the conductance of an ion pump is limited to one ion per absorbed photon of light, ion pumps never gained the same popularity.
In 2014, two groups independently reported6,8 genetically engineered chloride-conducting channelrhodopsins. Unlike ion pumps, these ion channels open to allow a large, rapid cellular influx of chloride anions in response to a single photon. These first-generation tools for optogenetics had only moderate sensitivity to light and poor temporal control, but were an excellent starting point from which improvements quickly followed.
Kato et al. now report the latest development: FLASH. This anion-conducting channelrhodopsin was designed using information gleaned from the two new crystal structures1,2, and supplants the previous best-in-class protein for inhibitory optogenetics17, ZipACR. FLASH can suppress individual action potentials in trains produced at frequencies of up to 40 hertz, with fewer off-target effects than ZipACR. It also inhibits neurons more reliably than ZipACR, reversibly reducing neuronal firing, on average, to about 30% of the initial rate in mouse brain slices. The authors show that when FLASH is expressed and activated in cells associated with the control of swimming in the nematode worm Caenorhabditis elegans, light almost completely inhibits swimming. We anticipate that any laboratory equipped for optogenetics should be able to use FLASH immediately.
But before FLASH can be widely adopted, proof is needed that its physiological side effects are minimal, and that it works in freely behaving fruit flies, zebrafish, mice and rats (four of the main animal models used by biologists). Moreover, temporal control of 40 Hz is probably acceptable for many applications, but 70% suppression of firing might not be sufficient for some experiments. And it remains to be seen whether long-term activation of the channel causes toxic effects in cells.
The current toolbox for inhibitory optogenetics is nowhere near as rich as that for excitatory experiments, but its expansion is following a similar timeline. We expect that the crystal structures reported by Kim et al. and Kato et al. will rapidly be used to engineer light-activated inhibitory channels that show substantial improvements over existing ones — FLASH represents a proof of concept of such efforts, rather than an end point. The new studies will surely inspire an array of tools to rival those of the excitatory family, preferably, but not limited to, proteins that: totally suppress action potentials; are ultrasensitive to light and ultrafast; and are activated by different wavelengths of light. By using both inhibitory and excitatory manipulations, and combining these with the latest in genetic tools, imaging technology, behavioural assays and computational modelling, our understanding of how our powerful brains function will deepen considerably.

 Read the paper: Crystal structure of the natural anion-conducting channelrhodopsin GtACR1
Read the paper: Crystal structure of the natural anion-conducting channelrhodopsin GtACR1
 Read the paper: Structural mechanisms of selectivity and gating in anion channelrhodopsins
Read the paper: Structural mechanisms of selectivity and gating in anion channelrhodopsins
 Ion channel in the spotlight
Ion channel in the spotlight
 Controlling neural circuits with light
Controlling neural circuits with light