- NEWS
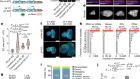
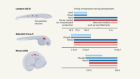
Tasmanian tiger genome offers clues to its extinction
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 552, 156-157 (2017)
doi: https://doi.org/10.1038/d41586-017-08368-1
References
Feigin, C. Y. et al. Nature Ecol. Evol. http://dx.doi.org/10.1038/s41559-017-0417-y (2017).
Miller, W. et al. Genome Res. 19, 213–220 (2009).
Murchison, E. P. et al. Cell 148, 780–791 (2012).