Abstract
Pancreatic ductal adenocarcinoma (PDAC) driven by oncogenic K-Ras remains among the most lethal human cancers despite recent advances in modern medicine. The pathogenesis of PDAC is partly attributable to intrinsic chromosome instability and extrinsic inflammation activation. However, the molecular link between these two events in pancreatic tumorigenesis has not yet been fully established. Here, we show that intracellular high mobility group box 1 (HMGB1) remarkably suppresses oncogenic K-Ras-driven pancreatic tumorigenesis by inhibiting chromosome instability-mediated pro-inflammatory nucleosome release. Conditional genetic ablation of either single or both alleles of HMGB1 in the pancreas renders mice extremely sensitive to oncogenic K-Ras-driven initiation of precursor lesions at birth, including pancreatic intraepithelial neoplasms, intraductal papillary mucinous neoplasms, and mucinous cystic neoplasms. Loss of HMGB1 in the pancreas is associated with oxidative DNA damage and chromosomal instability characterized by chromosome rearrangements and telomere abnormalities. These lead to inflammatory nucleosome release and propagate K-Ras-driven pancreatic tumorigenesis. Extracellular nucleosomes promote interleukin 6 (IL-6) secretion by infiltrating macrophages/neutrophils and enhance oncogenic K-Ras signaling activation in pancreatic lesions. Neutralizing antibodies to IL-6 or histone H3 or knockout of the receptor for advanced glycation end products all limit K-Ras signaling activation, prevent cancer development and metastasis/invasion, and prolong animal survival in Pdx1-Cre;K-RasG12D/+;Hmgb1−/− mice. Pharmacological inhibition of HMGB1 loss by glycyrrhizin limits oncogenic K-Ras-driven tumorigenesis in mice under inflammatory conditions. Diminished nuclear and total cellular expression of HMGB1 in PDAC patients correlates with poor overall survival, supporting intracellular HMGB1 as a novel tumor suppressor with prognostic and therapeutic relevance in PDAC.
Similar content being viewed by others
Introduction
Pancreatic ductal adenocarcinoma (PDAC) represents over 90% of all pancreatic malignancies. PDAC has the highest mortality rate of solid organ cancers, with only a 7% five-year survival rate1. PDAC shares some characteristics with other solid malignancies such as mutations affecting common signaling pathways, tumor heterogeneity, development of invasive malignancy from precursor lesions, inherited forms of the disease, and common environmental risk factors. However, unique obstacles have hindered the fight against PDAC. These include: 1) difficulties in diagnosis at an early stage in the disease because of the lack of specific symptoms or biomarkers and the anatomical location of the pancreas; 2) metastatic spread when the primary tumor is too small to detect using current methods; 3) dynamic interaction of the tumor with stromal cells, creating dense fibrous tissue around the tumor that contributes to therapeutic resistance; 4) avoidance of recognition and attack by the immune system, and 5) the small percentage of patients for whom curative surgery is a feasible option2,3. The need to further improve our understanding of pancreatic cancer biology, identify biomarkers, develop targeted therapies, and select eligible patient subgroups is unquestionably urgent.
Identification of clinically meaningful approaches relies heavily on the availability of preclinical models that recapitulate the key morphological and molecular features of the disease and offer high predictive value for clinically useful diagnostic and therapeutic interventions4,5. In particular, genetically engineered mouse models of PDAC have progressively improved in technical sophistication and, recapitulating features of human PDAC, and have had a measureable impact on our knowledge of tumorigenesis6,7,8. PDAC is characterized by a high frequency (> 95%) of activation of K-Ras mutations (especially G12D)9,10 and progresses from non-invasive pancreatic lesions that include pancreatic intraepithelial neoplasias (PanINs), intraductal papillary mucinous neoplasms (IPMNs), and mucinous cystic neoplasms (MCNs)11. Although oncogenic K-Ras plays a central role in controlling PDAC initiation and progression, the ability of mutant K-Ras to drive PDAC was not successfully investigated until the generation of mice with a Cre-inducible conditional allele (Pdx1-Cre;K-RasG12D/+, termed KC mice) targeting the endogenous K-Ras locus12. These KC mice develop lesions that slowly progress into advanced PDAC and have a median survival of 15 months12, suggesting that K-Ras activation is a tumor-initiating event that requires other elements that accelerate PDAC progression. Further understanding of K-Ras signaling and regulation may translate into improved treatments for pancreatic cancer.
High-mobility group box 1 (HMGB1) was first discovered as one of a group of chromatin-associated proteins with high acidic and basic amino acid content13. It is a highly-conserved protein with > 99% amino acid identity between murine and human molecules. Structurally, HMGB1 protein contains two homologous DNA-binding domains (termed A and B boxes, each 75 amino acids in length) with a negatively charged C-terminal region. Under normal conditions, most HMGB1 is localized in the nucleus to relax structural constraints within the nucleosome. Nuclear HMGB1 can bind to and bend DNA to control gene transcription, DNA repair, chromatin remodeling, and V(D)J recombination14,15. For example, HMGB1 is a transcriptional cofactor of p53, p73, the retinoblastoma protein, NF-κB, and nuclear hormone receptors including the estrogen receptor14,15. HMGB1 is also recognized as a damage-associated molecular pattern (DAMP) during cell death, inflammation, and encounter with various environmental stressors14,15,16,17. To act as a DAMP and danger signal, HMGB1 is required to be released by two different ways: active secretion from living immune cells or passive release from dead, dying, and injured cells. Dysfunction of intracellular and extracellular HMGB1 has been implicated in multiple human diseases or diverse pathologies including infections, cancer, neurodegeneration, aging, and heart disease18. In particular, HMGB1 was found to be a therapeutic target for tissue injury-mediated sterile inflammation and pathogen-mediated infection19. Interestingly, HMGB1 has dual roles in cancer development, progression, and therapy20,21. In many cases, extracellular HMGB1 acts as a pro-tumor protein due to its cytokine, chemokine, and growth factor activity, whereas intracellular HMGB1 promotes drug resistance due to its proautophagic activity22,23. However, the functional contribution of intracellular HMGB1 to tumorigenesis was previously unknown. In this study, we examined the impact of HMGB1 deficiency in pancreata on mutant K-Ras-driven initiation and progression of PDAC in mice, and investigated the underlying molecular mechanism as well as potential clinical significance. Our study indicates that intracellular HMGB1 is a novel tumor suppressor of PDAC by sustaining chromosome stability and limiting pro-inflammatory nucleosome release and activity.
Results
HMGB1 depletion accelerates initiation and progression of K-Ras-driven PDAC
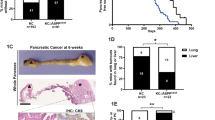
We created mice with conditional knockout of HMGB1 in the pancreas (Pdx1-Cre;Hmgb1−/−, termed CH mice) using Cre/loxP strategies and reported that CH mice exhibit normal pancreatic development and function under physiological conditions, but are more sensitive to experimental pancreatitis under pathological conditions24. Given that pancreatitis is considered one of the major risk factors for PDAC25, we postulated that HMGB1 would regulate K-Ras-driven pancreatic tumorigenesis and generated mice conditionally defective in both K-Ras and Hmgb1 in the pancreatic tissue (Pdx1-Cre;K-RasG12D/+;Hmgb1−/−, termed KCH mice; Supplementary information, Figure S1). These KCH mice were born at normal Mendelian frequency, exhibiting substantial weight loss and even sudden death at ∼3-6 weeks of age, resulting in a median survival of approximately three months (Figure 1A). These findings recapitulate those frequently found in humans with PDAC at presentation or progression. The average survival time of heterozygous HMGB1 knockout mice (Pdx1-Cre;K-RasG12D/+;Hmgb1+/−; termed KCH+/− mice) is slightly longer (4.9 months, Figure 1A), but with similar PDAC phenotypes observed in KCH mice (vide infra). These findings indicate that HMGB1 is involved in K-Ras-driven pancreatic cancer development.
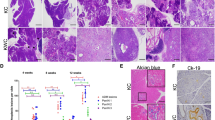
HMGB1 depletion accelerates initiation and progression of K-Ras-driven PDAC. (A) Kaplan-Meier survival analysis was performed in wild type (WT), CH (Pdx1-Cre;Hmgb1−/−), KC (Pdx1-Cre;K-RasG12D/+;Hmgb1+/+), KCH (Pdx1-Cre;K-RasG12D/+;Hmgb1−/−), and KCH+/− (Pdx1-Cre;K-RasG12D/+;Hmgb1+/−) mice (**P < 0.01, ***P < 0.001, log-rank test). (B) Incidence of pancreatic lesions, including pancreatic intraepithelial neoplasms (PanINs), intraductal papillary mucinous neoplasms (IPMNs), and mucinous cystic neoplasms (MCNs) in KC, KCH, and KCH+/− mice at indicated ages (n = 5 mice/genotype /age). (C) Percentages of ductal structures exhibiting normal morphology and indicated neoplastic ducts in KC, CH, KCH, and KCH+/− mice (n = 5 mice/genotype /age). (D) Representative histologic progression of pancreata in KC, CH, KCH, and KCH+/− mice at indicated ages shown by hematoxylin and eosin (H&E) staining (high resolution images shown in Supplementary information, Figure S2). (E) Percentages of intact acinar structures in pancreata from KC, CH, KCH, and KCH+/− mice at six weeks of age (n = 5 mice/genotype). (F) Incidence of PDAC in pancreata from KC, KCH, and KCH+/− mice (n = 5 mice/genotype/age). Representative samples H&E-stained for PDAC with stromal structure shown in right panel. (G) Incidence of tumor metastasis/invasion in KC, KCH, and KCH+/− mice at six to 24 weeks of age. Representative samples H&E-stained for tumor metastasis/invasion to the liver, lung, and kidney shown in upper panels. (H) Western blot analysis of HMGB1 expression in isolated tumor (T) or normal (N) tissue from KCH mice. (I) Cell proliferation analysis of indicated isolated tumor cells from KCH mice (n = 3, **P < 0.01, unpaired t-test).
We next assessed the impact of HMGB1 deletion in mice expressing K-RasG12D/+ on development of precursor lesions and PDAC. In line with a previous report12, control KC mice displayed no IPMNs or MCNs, only low grade PanINs (PanIN-1) at three months of age (Figure 1B and 1C) but high grade PanINs (PanIN-2 and -3) at nine months within an otherwise normal pancreas (data not shown). In contrast, pancreata from KCH mice exhibited 100% low and high grade PanINs at birth (Figure 1B and 1C), occupying the entire pancreas and resulting in almost complete loss of normal pancreatic tissue as early as day 7 (Figure 1D and Supplementary information, Figure S2). KCH+/− mice also exhibited low and high grade PanINs on day 3 (Figure 1B-D). In addition to PanINs, cystic lesions, including IPMNs and MCNs, also appeared in pancreata from KCH and KCH+/− mice (Figure 1B-1D and Supplementary information, Figure S3), as frequently observed in human patients26. The percentage of normal pancreatic acinar cells was remarkably reduced in KCH and KCH+/− mice (Figure 1D and 1E and Supplementary information, Figure S2) with decreased exocrine function (Supplementary information, Figure S4A and S4B). Blood glucose levels are tightly controlled by regulation of insulin release from pancreatic beta-cells. We observed that blood glucose levels were increased (Supplementary information, Figure S4C) whereas serum insulin levels were decreased (Supplementary information, Figure S4D) in KCH mice due to the loss of pancreatic islet beta-cells (Supplementary information, Figure S4E and S4F). Unlike KCH mice, CH mice had normal exocrine and endocrine function, as demonstrated in our previous study24. These findings indicate that loss of HMGB1 is dispensable for normal pancreatic development, but promotes pancreatic function insufficiency during K-Ras-driven tumor development.
It is clear that pancreatic lesions are mainly derived from acinar cells undergoing acinar to ductal metaplasia (ADM), an event usually induced by pancreatitis27. We therefore hypothesized that loss of HMGB1 may promote ADM in K-Ras-driven pancreatic tumorigenesis. Indeed, KCH and KCH+/− mice exhibited significant ADM, characterized by colocalization of amylase (an acinar cell marker) and cytokeratin 19 (CK19, a ductal cell marker) within lesions (Supplementary information, Figure S5A). The expression of sex determining region Y-box 9 (SOX9), a core regulator of ADM and initiation of PDAC, has been identified in PanINs, IPMNs, and MCNs in mouse and human pancreatic cancer specimens28. Consistently, the expression of SOX9 was elevated in KCH and KCH+/− mice (Supplementary information, Figure S5B). Thus, loss of HMGB1 is an accelerating event for the development of these various premalignant neoplasms (e.g., ADM and SOX9 expression) of PDAC.
We explored further the role of HMGB1 in PDAC development. In KC mice, the onset of PDAC usually occurs beyond 12 months12. In contrast, KCH mice between six and 24 weeks old exhibited progression to either locally invasive PDAC (Figure 1F and Supplementary information, Figure S3) or PDAC with increased metastatic or invasion capacity to the liver, lung, and kidney (Figure 1G and Supplementary information, Table S1). Tumors in the liver (but not lung or kidney) shared a similar morphology to pancreatic tumor in KCH mice (Figure 1G). Western blot analysis confirmed loss of HMGB1 protein expression in metastatic or invading tumor from liver, lung, and kidney (Figure 1H). Moreover, increased cell proliferation was observed in isolated metastatic or invading tumor cells from lung (but not liver or kidney) compared to HMGB1-deficient primary pancreatic tumor cells (Figure 1I). Further characterization of KCH and KCH+/− mice with immunofluorescence microscopy of pancreata showed that the loss of HMGB1 increased ductal lesions, as determined by CK19 positivity (Supplementary information, Figure S6A) and mucin expression based on Alcian blue staining (Supplementary information, Figure S6B). Notably, pancreata from KCH and KCH+/− mice also showed abundant stromal components (a hallmark of PDAC), as determined by vimentin positivity (Supplementary information, Figure S7A) and Masson's trichrome staining (Supplementary information, Figure S7B). Ki-67 (Supplementary information, Figure S8A) staining of the ductal epithelium showed that loss of HMGB1 enhanced K-Ras-induced cellular proliferation. Consistent with the increased matrix metalloproteinase-7 (MMP7) expression observed in pre-invasive and invasive human pancreatic cancer specimens29, we also found elevated expression of MMP7 in ductal lesions of KCH and KCH+/− mice (Supplementary information, Figure S8B). Collectively, this changed histology confirmed that loss of HMGB1 accelerates K-Ras-driven pancreatic tumorigenesis and development.
Oxidative DNA damage promotes pancreatic tumorigenesis in KCH mice
Chromosomal instability is an intrinsic pathway responsible for mutagenesis and carcinogenesis30. Despite its widespread prevalence, knowledge of the mechanisms and contributions of chromosomal instability in cancer remains largely unknown. As HMGB1 acts as a DNA chaperone, influencing multiple processes in chromatin such as gene transcription and nucleosome assembly31,32, we postulated that HMGB1 regulates chromosomal stability and the DNA damage response in pancreatic tumorigenesis. Telomere analysis by fluorescence in situ hybridization showed an increase in morphological defects (including chromosome fusion and concatenation, apparent loss of telomeres, and sister telomere fusion) in ductal cells from KCH mice compared with KC mice (Figure 2A). Levels of histone H2AX phosphorylation (γ-H2AX, a marker for DNA damage33; Figure 2B) and 8-hydroxyguanine (8-OHDG, a marker of oxidative DNA damage34; Figure 2C) were also upregulated in the ductal lesions of KCH mice. These were accompanied by abnormal expression of genes involved in regulating the DNA damage response (Supplementary information, Figure S9A) and telomere maintenance (Supplementary information, Figure S9B) in KCH mice compared with KC mice. Thus, loss of HMGB1 in the pancreas accelerates oxidative DNA damage and chromosome instability in K-Ras-driven pancreatic tumorigenesis.
Oxidative DNA damage promotes pancreatic tumorigenesis in KCH mice. (A) Percentage of abnormal telomeres in ductal cells from KC and KCH mice at six weeks of age (n = 5 mice/genotype). CF = chromosome fusions and concatenation; SFE = telomere signal-free end; STF = sister telomere fusion. Total = CF + SFE + STF. (B-C) Immunohistochemical staining of DNA damage marker (γ-H2AX) and oxidative DNA damage marker (8-OHDG) in KC and KCH mice. (D) ELISA analysis of serum nucleosome levels in KC and KCH mice (n = 5 mice/genotype/age). (E-I) Antioxidant N-acetylcysteine (NAC, 300 mg/kg i.p., five times per week, started at 4 weeks of age for four weeks) treatment prolonged survival in KCH mice (E, **P < 0.01, log-rank test) with decreased telomere defects (F), histone H2AX phosphorylation (G), oxidative DNA damage (H), and serum nucleosome levels (I). *P < 0.05, **P< 0.01 (n = 5 mice/genotype, unpaired t-test).
Pancreatitis is an extrinsic pathway contributing to the pathogenesis of PDAC, and inflammatory mediators exert pleiotropic effects in the development of PDAC35. Extracellular nucleosomes including histone and DNA have been suggested to be inflammatory mediators for cell death36. We previously demonstrated that loss of HMGB1 in the pancreas increases pro-inflammatory nucleosome release in experimental pancreatitis by oxidative injury24. Accordingly, serum nucleosome levels were elevated in KCH mice compared with KC mice at 1 to 24 weeks of age (Figure 2D). Furthermore, we found that treating 4-week-old KCH mice with the antioxidant N-acetylcysteine (NAC) for 4 weeks significantly prolonged animal survival (Figure 2E) with decreased telomere defects (Figure 2F), histone H2AX phosphorylation (Figure 2G), oxidative DNA damage (Figure 2H) and serum nucleosome levels (Figure 2I). These findings suggest that oxidative DNA damage-mediated nucleosome release contributes to PDAC in KCH mice.
Extracellular histone promotes pancreatic tumorigenesis in KCH mice
Next, we investigated whether suppression of extracellular nucleosome activity could impact PDAC development in KCH mice. As histone H3 is one of the five main histone proteins in the nucleosome, we tested the effects of neutralizing blood nucleosome activity with an anti-H3 antibody upon tumor development in KCH mice. Notably, this treatment resulted in prolonged animal survival (Figure 3A), decreased pancreatic lesion formation (Figure 3B), increased normal acinar structures (Figure 3B), and reduced metastasis/invasion to the liver, lung, and kidney (Figure 3C). These findings indicate that circulating nucleosomes promote pancreatic tumorigenesis in KCH mice.
Extracellular histone promotes pancreatic tumorigenesis in KCH mice. (A) H3 Ab or IL-6 Ab treatment (10 mg/kg i.p., twice per week, started at four weeks of age for four weeks) prolonged survival in KCH mice at 12 weeks of age (n = 10 mice/treatment, **P < 0.01, ***P < 0.001, log-rank test). The median survival of untreated, IgG-treated, H3 Ab-treated, and IL-6 treated KCH mice were nine, nine, 12, and 12 weeks, respectively, in this setting. (B-I) In parallel, pancreatic lesion formation (B), incidence of tumor metastasis/invasion (C), serum IL-6 level (D), percentage of tumor infiltration of macrophages (E) and neutrophils (F), and relative expression of p-ERK1/2 (G), p-AKT (H), and p-STAT3 (I) in the pancreas were assayed. Graphs show means ± sem, *P < 0.05, **P < 0.01, ***P < 0.001 (n = 5 mice/treatment, unpaired t-test).
We and others have previously reported that interleukin (IL-6) is a key PDAC-associated proinflammatory cytokine promoting K-Ras and activating the STAT3 (signal transducer and activator of transcription 3) signaling in PDAC37,38,39,40,41. Constitutive activation of STAT3 is implicated in stem cell self-renewal, cancer cell survival, and inflammation in PDAC. We next sought to determine whether the IL-6-STAT3 pathway is affected by circulating nucleosome in KCH mice. We found that the serum IL-6 level (Figure 3D) was also reduced in KCH mice following treatment with anti-H3 antibody. Analysis of tumor infiltrations of myeloid cells such as macrophages (Figure 3E) and neutrophils (Figure 3F) by staining for F4/80 or Gr-1 antibody, respectively, revealed that these major cellular sources of IL-6 were decreased in KCH mice following treatment with anti-H3 antibody.
Further characterization of the activation of phospho-extracellular signal-regulated kinase-1/2 (p-ERK1/2) (Figure 3G), phospho-V-Akt murine thymoma viral oncogene homolog (p-AKT) (Figure 3H), and p-STAT3 (Figure 3I) in the pancreas revealed striking differences following anti-H3 antibody treatment. The increased levels of p-ERK1/2 (Figure 3G), p-AKT (Figure 3H), and p-STAT3 (Figure 3I) in the pancreata (especially tumor lesions) of KCH mice were suppressed by treatment with anti-H3 antibody. Accordingly, treatment with anti-IL-6-neutralizing antibody also increased animal survival (Figure 3A) and decreased levels of p-ERK1/2 (Figure 3G), p-AKT (Figure 3H), and p-STAT3 (Figure 3I) in the pancreata of KCH mice. These findings suggest that extracellular nucleosome activity contributes to the inflammatory response, sustaining IL-6/STAT3 signaling within the pancreatic tumor microenvironment.
Deletion of RAGE protects against pancreatic tumorigenesis in KCH mice
Several receptors, including the receptor for advanced glycation end products (RAGE)42, Toll like receptor (TLR)-243, TLR443, and TLR944, mediate the biological activity of extracellular nuclear DAMP in individual cell types. Knockout of RAGE or TLR9 inhibits development of pancreatic lesions and progression to PDAC in KC mice39,45. Expression of protein (Supplementary information, Figure S10A and S10B) and mRNA (Supplementary information, Figure S10C) of RAGE and TLR9 (but not TLR2 and TLR4) was increased in the pancreas of KCH mice compared with KC mice, indicating a potential role of RAGE and TLR9 in pancreatic tumorigenesis.
To identify whether RAGE or TLR9 is required for PDAC development in KCH mice, we crossed KCH mice with Rage−/− or Tlr9−/− mice to generate KCHR (Pdx1-Cre;K-RasG12D/+;Hmgb1−/−;Rage−/−) and KCHT9 (Pdx1-Cre;K-RasG12D/+;Hmgb1−/−;Tlr9−/−) mice. Notably, deletion of RAGE (but not TLR9) significantly prolonged animal survival (Figure 4A), decreased formation of pancreatic lesions (Figure 4B), increased normal acinar structures (Figure 4B), limited tumor metastasis/invasion to the liver, lung, and kidney (Figure 4C), reduced serum IL-6 levels (Figure 4D), and diminished pancreatic levels of p-ERK1/2 (Figure 4E), p-AKT (Figure 4F) and p-STAT3 (Figure 4G) in KCHR mice compared with KCH mice. These findings suggest that RAGE (but not TLR9) is required for oncogenic K-Ras activation in KCH tumor development.
Deletion of RAGE protects against pancreatic tumorigenesis in KCH mice. (A) Kaplan-Meier survival analysis was performed in KCH (Pdx1-Cre;K-RasG12D/+;Hmgb1−/−), KCHR (Pdx1-Cre;K-RasG12D/+;Hmgb1−/−;Rage−/−), and KCHT9 (Pdx1-Cre;K-RasG12D/+;Hmgb1−/−;Tlr9−/−) mice (n = 5 mice/treatment, **P < 0.01, log-rank test). (B-G) In parallel, pancreatic lesion formation (B), tumor metastasis/invasion incidence (C), serum IL-6 level (D), and relative expressions of p-ERK1/2 (E), p-AKT (F), and p-STAT3 (G) in the pancreas were assayed. Graphs show means ± sem, *P < 0.05, **P < 0.01 (n = 5 mice/treatment, unpaired t-test).
Moreover, KCHR mice (but not KCHT9 mice) exhibited decreased pancreatic expression of SOX9 (an ADM marker), vimentin (a stromal marker), Ki-67 (a proliferation marker), and MMP7 (an invasion marker) compared with KCH mice (Supplementary information, Figure S11). We extended our findings with cultured peritoneal macrophages and PDAC cells from KCH, KCHR, and KCHT9 mice. We found that loss of RAGE (but not TLR9) diminished recombinant nucleosome-induced IL-6 release in isolated peritoneal macrophages and PDAC cells in vitro (Supplementary information, Figure S12). Together, these data suggest that RAGE is required for K-Ras-driven tumorigenesis and development partly through mediating extracellular nucleosome activity in KCH mice.
Pharmacologic inhibition of nuclear HMGB1 loss and release limits K-Ras-induced pancreatic lesions
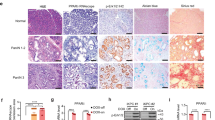
We next investigated the expression and release of HMGB1 in K-RasG12D-induced oncogenic transformation. Intracellular HMGB1 (especially nuclear HMGB1) expression was progressively reduced over 72h (Figure 5A), whereas extracellular HMGB1 progressively increased over the same interval (Figure 5B) in response to K-RasG12D-induced oncogenic transformation of normal pancreatic acinar cells. This suggests that HMGB1 loss and release is an important event in pancreatic tumorigenesis. Several compounds have been identified that directly or indirectly attenuate the translocation and release of HMGB1. Among these compounds, glycyrrhizin is a direct HMGB1 inhibitor with anti-inflammatory and antioxidant activity46,47. We therefore examined the effect of pharmacological inhibition of HMGB1 by glycyrrhizin in K-RasG12D-induced oncogenic transformation. We found that glycyrrhizin treatment led to a remarkable suppression of nuclear loss (Figure 5C) and extracellular release of HMGB1 (Figure 5D) in K-RasG12D-induced oncogenic transformation. Moreover, glycyrrhizin also blocked K-RasG12D-induced increase in p-AKT and p-ERK1/2 (Figure 5C), suggesting that pharmacologic inhibition of nuclear HMGB1 loss and release can limit activation of K-Ras signaling.
Inhibition of nuclear HMGB1 loss and release by glycyrrhizin limits K-Ras-induced PanIN formation. (A, B) Effects of K-RasG12D transfection on HMGB1 expression (A) and release (B) in primary mouse pancreatic acinar cells (n = 3, ***P < 0.001, data are expressed as means ± sem, unpaired t-test). (C, D) Effects of glycyrrhizin (500 μM) on HMGB1 expression (C) and release (D) in primary mouse pancreatic acinar cells after K-RasG12D transfection for 48 h (n = 3, ***P < 0.001, data are expressed as means ± sem, unpaired t-test). (E-G) Effects of glycyrrhizin (10 mg/kg) on PanIN formation (E) and HMGB1 expression (F) and release (G) on day 21 in a cerulein-mediated accelerated oncogenic K-Ras mouse model of pancreatic cancer (n = 5 mice/treatment, ***P < 0.001, data are expressed as means ± sem, unpaired t-test).
It is well established that cerulein treatment-induced pancreatitis accelerates PanIN formation and stromal response in KC mice48,49. Next, we determined whether pharmacologic inhibition of nuclear HMGB1 loss and release by glycyrrhizin prevents K-Ras-induced pancreatic cancer initiation under inflammatory conditions. Indeed, we found that glycyrrhizin treatment prevented PanIN development preserving normal acinar structures (Figure 5E), sustaining nuclear HMGB1 expression (Figure 5F), and decreasing circulating HMGB1 and nucleosome levels (Figure 5G) in the cerulein-mediated accelerated oncogenic K-Ras mouse model of pancreatic cancer. Hence, prevention of loss of nuclear HMGB1 and subsequent nuclear DAMP (e.g., nucleosome and HMGB1) release may reveal a new PDAC prevention strategy.
Reduced HMGB1 expression correlates with poor survival in pancreatic cancer patients
To determine whether HMGB1 expression is aberrant in human pancreatic cancer, we used pancreas tissue microarrays to analyze a cohort of 90 patients with pancreatic cancer. The expression of HMGB1 protein, especially nuclear HMGB1, was decreased in pancreatic tumor compared with adjacent normal pancreatic tissue and normal tissue (Figure 6A). Furthermore, the proportion of PDAC tumors that had lower HMGB1 expression than normal adjacent tissue (low HMGB1 group) correlated with a worse outcome (Figure 6B). The median survival time of the low HMGB1 group versus the high group was 10 months versus 43 months. However, HMGB1 mutation was not observed in pancreatic cancer patients according to the cBioPortal website (http://www.cbioportal.org). These findings indicated that loss of HMGB1 expression (but not HMGB1 mutation) may be a pathological event in human PDAC progression.
Discussion
The pathogenesis of PDAC is partly attributable to intrinsic chromosome instability and extrinsic inflammation activation25,50,51. However, the molecular link between these two events in pancreatic tumorigenesis has not yet been fully established. We show that intracellular HMGB1 confers remarkable suppression of oncogenic K-Ras-driven pancreatic tumorigenesis by inhibiting pro-inflammatory nucleosome release mediated by chromosome instability characterized by chromosome rearrangements and telomeric defects (Figure 7). Conditional genetic ablation of either single or both alleles of HMGB1 in the pancreas renders mice extremely sensitive to oncogenic K-Ras-driven initiation of precursor lesions at birth, as well as tumor metastasis/invasion at six weeks. Loss of HMGB1 in the pancreas enables oxidative DNA damage and emergent chromosomal instability, which leads to inflammatory nucleosome release and propagates K-Ras-driven pancreatic tumorigenesis. Extracellular nucleosomes promote IL-6 secretion by infiltrating macrophages/neutrophils and sustain oncogenic K-Ras signaling activation in pancreatic lesions. Neutralizing antibodies to IL-6 or histone H3 or knockout of RAGE all limit K-Ras signaling activation, prevent cancer development and metastasis/invasion, and prolong animal survival in KCH mice. Pharmacological inhibition of intracellular HMGB1 loss limits K-Ras-driven tumorigenesis in mice. This evidence strongly indicates that intracellular HMGB1 is a tumor suppressor of PDAC.
Schematic depicting HMGB1 deficiency-mediated nucleosome release linking chromosomal instability to the inflammatory response in K-Ras-driven PDAC. Oncogenic K-RasG12D leads to HMGB1 translocation and release into the extracellular space. Loss of intracellular HMGB1 increases chromosomal instability and promotes nucleosome release (current study). In addition, we previously demonstrated that extracellular nucleosome activates innate immune cells (e.g., macrophages) to secrete HMGB1 into circulation24. Both extracellular nucleosome and HMGB1 exacerbates PDAC development by enhancing RAGE-dependent inflammatory responses.
HMGB1 was initially defined in 1973 as a non-histone chromatin protein. Inside the nucleus, HMGB1 interacts with DNA and histones to determine chromatin structure32 and regulates key processes such as transcription52. HMGB1 has other intracellular actions; when it translocates to the cytoplasm, it can mediate autophagy53,54. Outside the cell, HMGB1 acquires a new identity to serve as a danger signal or a DAMP55. Elevated serum HMGB1 levels are implicated in disease with sterile inflammation and infection19. Elevated serum HMGB1 levels also correlate with the presence, progression, and prognosis of PDAC56. Although serum levels are increased in PDAC patients, the expression of intracellular HMGB1 in pancreatic tissue remains unclear. Our tissue microarray analysis of patients with pancreatic cancer reveals a significant correlation between reduced HMGB1 expression and poor survival. We also observed that K-RasG12D-induced oncogenic transformation led to HMGB1 translocation and release into the extracellular space. On one hand, loss of intracellular HMGB1 increases chromosomal instability and telomere attrition, whereas increased extracellular nuclear DAMPs (e.g., nucleosome) in HMGB1-deficient cancer cells cause an excessive inflammation response in the tumor microenvironment. These HMGB1-mediated pathogenic changes drive the initiation and development of PDAC.
Telomeres are the protective DNA-protein complexes found at the end of eukaryotic chromosomes, whereas subtelomeres are segments of DNA between telomeric caps and chromatin57. Previous studies showed that knockout of the HMGB1 in mouse embryonic fibroblasts or knockdown of HMGB1 in human breast cancer cell lines resulted in a decline in telomerase activity and telomere dysfunction58,59. We found here that loss of HMGB1 leads to abnormal telomeres and nucleosome release in PDAC cells. It will be interesting to uncover whether HMGB1-mediated release of nucleosome is subtelomere- or telomere-specific.
Metaplasia is defined by the conversion or replacement of one differentiated cell type with another in the context of a given tissue. In some tissues, metaplasia is associated with an increased risk of cancer60. Investigation of metaplasia might also provide clues to the cell of cancer origin. To that end, pancreatic acinar cells have the capacity to undergo metaplasia to a ductal cell phenotype in the origin of PDAC61,62,63,64. ADM might represent reprogramming of a progenitor population, direct transdifferentiation of acinar cells to ductal cells, or transdifferentiation via an intermediate cell type (potentially a progenitor cell)65. We demonstrate that loss of HMGB1 increases K-Ras-driven ADM in pancreatic tumorigenesis. The molecular basis underlying the development of ADM involves a number of seemingly diverse pathways and molecules, including transcriptional factor65,66,67,68. Sox9 is a critical transcriptional factor of the HMG box family, which not only sustains proliferation and survival of the pancreatic progenitor cell pool69,70, but also triggers ADM in neoplastic transformation including PanINs, IPMNs, and MCNs28,63,71,72,73. We observed that loss of HMGB1 increased PanINs, IPMNs, and MCNs, which is associated with increased Sox9 expression in K-Ras-driven pancreatic tumorigenesis. The detailed molecular basis for HMGB1 function in the formation of ADM and mixed precursor lesions warrants further investigation.
Growing evidence shows that tumors, including PDAC, are sustained and promoted by inflammatory signals from the surrounding microenvironment. Our current study identifies a role of nucleosome release in the pancreatic tumor microenvironment. Besides exerting intranuclear functions, nucleosomes act as inflammatory DAMPs when they are released into the extracellular space in oxidative injury and genomic instability74. Indeed, increased serum levels of nucleosomes, including the component histones and DNA, have been observed in many cancers and correlate with disease progression75,76. As nucleosomes are stable structures in the circulation77, they could be a valuable source of biomarkers in PDAC. Our current study provides evidence that extracellular nucleosomes promote IL-6 secretion by myeloid cells, which in turn sustains STAT3 and K-Ras signaling in pancreatic cancer cells. These findings may represent a new response of the inflammatory microenvironment in tumor progression.
Both the present and previous studies highlight the important role of RAGE in the initiation of PDAC. RAGE is a transmembrane receptor of the immunoglobulin gene superfamily and a multifunctional receptor within the tumor microenvironment20. RAGE and its ligands are linked to the development and progression of several cancers by facilitating the maintenance of a chronic inflammatory state78 and/or by promotion of metastases22. We previously demonstrated that: (1) RAGE was highly expressed in mouse and human PDAC39; (2) Targeted genetic ablation of RAGE in mice prevented experimental acute pancreatitis79 and pancreatic cancer growth in the KC model39; and (3) RAGE was essential for pancreatitis79 and oncogenic K-Ras-mediated hypoxic signaling in pancreatic cancer development80. Several studies have found that RAGE is a nucleic acid receptor for recognition of various DNA and RNA molecules42,81. Our recent study shows that RAGE is a potential receptor for nuclear DAMP complex (HMGB1-histone-DNA) in macrophages82. Nucleosomes may signal through receptors such as TLRs, as well as other yet unidentified receptors, depending on cell type36. In the present study, we further identified RAGE (but not TLR9) as a receptor mediating extracellular nucleosome activity in K-Ras-driven tumorigenesis in KCH mice. Pharmacological or genetic blocking of the nucleosome-RAGE pathway limits STAT3 and K-Ras signaling in KCH mice.
In summary, conditional knockout of HMGB1 in the pancreas leads to significant acceleration of K-Ras-driven carcinogenesis. Compared with other reported genetically engineered mouse models, the KCH model is extremely sensitive to oncogenic K-Ras signaling, with development of mixed precursor lesions at birth. HMGB1 translocation from the nucleus or deficiency-mediated nucleosome release can now be appreciated as key events linking chromosomal instability and the inflammatory response (Figure 7). This process requires activation of RAGE, a DAMP receptor that promotes inflammatory responses to HMGB183, DNA42, and histones79. Thus, an impaired HMGB1-nucleosome-RAGE-mediated DAMP pathway contributes to K-Ras signaling activation and may represent a potent therapeutic target in PDAC. The KCH model provides an interesting and provocative model for future translational research and drug discovery.
Materials and Methods
Animals and in vivo models
We conducted all animal care and experiments in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care guidelines (http://www.aaalac.org) and with approval from the University of Pittsburgh Institutional Animal Care and Use Committee. C57BL/6 mice were purchased from Jackson Laboratories.
Hmgb1flox/flox mice on C57BL/6 background were obtained from Dr Eugene B Chang (University of Chicago)24,84. The HMGB1 conditional knockout mice were genotyped with primers 5′-TGATGCGAACACGGCGTGCTCTA-3′ and 5′- GCACAAAGAATGCATATGAGGAC-3′. PCR conditions used were as follows: 94 °C for two minutes; then 30 cycles of 94 °C for 15 s, 62 °C for 30 s and 72 °C for 30 s; and then held at 72 °C for 5 min before cooling to 4 °C. WT mice were identified by a single band at 635 bp. HMGB1 floxed mice were identified by a single band at 700 bp.
Rage−/− mice on C57BL/6 background were a kind gift from the late Dr Angelika Bierhaus (University of Heidelberg)85. Dr Bierhaus passed away on 15 April 2012 after a long and courageous battle with cancer. The RAGE knockout mice were genotyped with primers 5-CCTGGGTGCTGGTTCTTG-3′ and 5′- CTGAGGTCCGTGGCTAGG-3′. PCR conditions used were as follows: 94 °C for five minutes; then 30 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 3 min; and then held at 72 °C for 5 minutes before cooling to 4 °C. Rage+/+ mice were identified by a single band at 1 750 bp. Rage−/− mice were identified by a single band at 2 100 bp.
Tlr9−/− mice lacked CpG motif on C57BL/6 background was obtained from Dr Timothy R Billiar (University of Pittsburgh)86. The Tlr9−/− mice had a one base-pair mutation on CpG. The section of DNA for sequencing was ACGCCCCCAGT. The CCCCC 5 C's were a Tlr9−/− and CCTCC were a Tlr9+/+.
Pdx-1-Cre and K-rasG12D/+ transgenic mice on C57BL/6 background were received from the MMHCC/NCI Mouse Repository. These mice were crossed to generate indicated KC, KCH, KCHR, and KCHT9 animals. All mice were housed under a 12-h light-dark diurnal cycle with controlled temperature (21 °C to 23 °C) and provided with a standard rodent diet and water ad libitum throughout all experiments.
To study the effects of oxidative DNA damage on pancreatic tumorigenesis, four-week-old KCH mice were injected intraperitoneally (i.p.) with NAC (10 mg/kg, Sigma) five times per week for four weeks. To study the effects of extracellular nucleosome on pancreatic tumorigenesis, four-week-old KCH mice were injected i.p. with neutralizing anti-H3 (10 mg/kg, Abcam) or anti-IL-6 antibody (10 mg/kg, BioLegend) twice per week for 4 weeks.
To study the effects of HMGB1 inhibitor on pancreatitis-induced pancreatic tumorigenesis, four-week-old KC mice were treated with two sets of six hourly i.p. cerulein injections (50 μg/kg, Sigma) on alternating days separated by 24 h. These KC mice were then injected i.p. with glycyrrhizin (10 mg/kg, Sigma) five times per week for 2 weeks. The final day of cerulein injection was considered day 0.
ELISA analysis
The amylase (Abcam), trypsin (Abcam), IL-6 (BioLegend), insulin (Thermo Fisher Scientific Inc.), and HMGB1 (Shino-Test Corporation) levels in serum or supernatant fractions from pancreatic homogenate were measured using ELISA according to the manufacturer's protocol. For measurement of serum nucleosomes, Cell Death Detection ELISAplus (Roche Diagnostics) was used. It is based on a quantitative sandwich enzyme immunoassay principle. Monoclonal mouse antibodies directed against DNA (single-stranded and double-stranded) and histones (H1, H2A, H2B, H3, and H4) specifically detect mono- and oligonucleosomes. The anti-histone antibody additionally binds to the microtiter plate, whereas the anti-DNA antibody labeled with peroxidase reacts with 2,2′-azinobis(3-ethylbenzothiazoline sulfonate)87. Recombinant nucleosomes were purchased from EpiCypher.
Image analysis
For histological analysis, pancreatic specimens were fixed with 10% buffered formalin, dehydrated in ethanol, embedded with paraffin, and stained with hematoxylin and eosin, Alcian blue, or Masson's trichrome. The fraction of preserved acinar area was calculated as previously described88. Pancreatic ductal dysplasia was graded according to established criteria89. Histological images were acquired using an EVOS FL Auto Cell Imaging System (Thermo Fisher Scientific, Inc.).
Immunofluorescent staining of mouse tissues was performed using antibodies directed against 8-OHDG (Abcam), γ-H2AX (Cell Signal Technology), p-ERK1/2 (Cell Signal Technology), p-AKT (Cell Signal Technology), p-STAT3 (Cell Signal Technology), amylase (Santa Cruz Biotechnology), CK19 (Developmental Studies Hybridoma Bank), SOX9 (Millipore), vimentin (Cell Signal Technology), ki67 (Abcam), MMP7 (Cell Signal Technology), RAGE (Santa Cruz Biotechnology), TLR2 (Santa Cruz Biotechnology), TLR4 (Abcam), TLR9 (Abcam), insulin (Cell Signal Technology), glucagon (Abcam), and DAPI (Invitrogen). Immunofluorescent images were acquired using an AxioObserver Z1 Microscope with Apotome (Carl Zeiss). Quantifications of images were performed by assessing 20X high-power fields per slide and 20 fields per mouse (five mice/group) in a blinded manner.
Immunohistochemistry in human pancreatic cancer tissue array was performed using antibodies directed against HMGB1 (Abcam). This tissue microarray (#HPan-Ade180Sur-01) was purchased from US Biomax and included 90 cases of pancreatic cancer tumor (one core/case) and matched normal adjacent tissue (one core/case). Clinical stage, survival information, smoking history, drinking history, diabetes history, hepatitis B information, and family medical history were available from the US Biomax website (http://www.biomax.us/tissue-arrays/Pancreas/HPan-Ade180Sur-01).
Quantitative real-time PCR
Total RNA was extracted using TRI reagent (Sigma) according to the manufacturer's instructions. First-strand cDNA was synthesized from 1 μg of RNA using the iScript cDNA Synthesis kit (Bio-Rad, Hercules, CA). cDNA from various cell samples were amplified using real-time quantitative PCR with specific primers (Rage: 5′-CTGCGTGTTCTACGAGATTGCC-3′ and 5′-GACAAGGTGTGCCGATGACATC-3′; Tlr2: 5′- ACAGCAAGGTCTTCCTGGTTCC-3′ and 5′- GCTCCCTTACAGGCTGAGTTCT-3′; TLR4: 5′- AGCTTCTCCAATTTTTCAGAACTTC-3′ and 5′-TGAGAGGTGGTGTAAGCCATGC-3′; Tlr9: 5′- GCTGTCAATGGCTCTCAGTTCC-3′ and 5′- CCTGCAACTGTGGTAGCTCACT-3′) and the data were normalized to Rn18s ribosomal RNA (5′-CTTAGAGGGACAAGTGGCG-3′ and 5′-ACGCTGAGCCAGTCAGTGTA-3′). RT² profiler PCR array for telomeres or DNA damage signaling pathway was measured using a Kit from QIAGEN according to the manufacturer's protocol.
K-RasG12D transfection
K-RasG12D plasmid (Addgene) was electroporated into primary mouse acinar cells using the Neon Transfection System (Life technologies) according to the manufacturer's protocol.
Measuring glucose
Glucose concentration measurements were obtained from whole-blood samples using hand-held whole-blood glucose monitors (Bayer) according to the manufacturer's protocol.
Western blot analysis
Western blot was used to analyze protein expression as described previously54. In brief, after extraction, proteins in cell lysates were first resolved by SDS-PAGE electrophoresis and then transferred to nitrocellulose membrane and subsequently incubated with primary antibodies (anti-HMGB1 (Novus), anti-actin (Sigma), anti-GAPDH (Sigma), anti-fibrillarin (Abcam), anti-AKT (Cell Signal Technology), anti-p-AKT (Cell Signal Technology), anti-ERK1/2 (Cell Signal Technology), and anti-p-ERK1/2 (Cell Signal Technology)). After incubation with peroxidase-conjugated secondary antibodies, the signals were visualized with enhanced chemiluminescence (Pierce, Rockford, IL, USA) according to the manufacturer's instructions.
Statistical analysis
Data are presented as mean ± sem. Survival was measured using the Kaplan–Meier method. Statistical significance was determined with unpaired t-test or the log-rank test using SigmaPlot 11.0 software. P < 0.05 was considered significant.
Author Contributions
RK and DT designed the research. RK, YX, QZ, WH, QJ, ZS, DZ, and DT performed the experiments. RK, HW, HJZ, MTL, and DT analyzed the results. JB, TRB, MTL, and HJZ provided important reagents. RK, HW, and DT wrote the paper. JB, TRB, DLB, MTL, and HJZ edited and commented on the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
Siegel RL, Miller KD, Jemal A . Cancer statistics, 2015. CA Cancer J Clin 2015; 65:5–29.
Ryan DP, Hong TS, Bardeesy N . Pancreatic adenocarcinoma. N Engl J Med 2014; 371:1039–1049.
Garrido-Laguna I, Hidalgo M . Pancreatic cancer: from state-of-the-art treatments to promising novel therapies. Nat Rev Clin Oncol 2015; 12:319–334.
Cook N, Jodrell DI, Tuveson DA . Predictive in vivo animal models and translation to clinical trials. Drug Discov Today 2012; 17:253–260.
Boj SF, Hwang CI, Baker LA, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 2015; 160:324–338.
Mazur PK, Siveke JT . Genetically engineered mouse models of pancreatic cancer: unravelling tumour biology and progressing translational oncology. Gut 2012; 61:1488–1500.
Saluja AK, Dudeja V . Relevance of animal models of pancreatic cancer and pancreatitis to human disease. Gastroenterology 2013; 144:1194–1198.
Perez-Mancera PA, Guerra C, Barbacid M, Tuveson DA . What we have learned about pancreatic cancer from mouse models. Gastroenterology 2012; 142:1079–1092.
Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ . Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov 2014; 13:828–851.
Stephen AG, Esposito D, Bagni RK, McCormick F . Dragging ras back in the ring. Cancer Cell 2014; 25:272–281.
Hruban RH, Adsay NV . Molecular classification of neoplasms of the pancreas. Hum Pathol 2009; 40:612–623.
Hingorani SR, Petricoin EF, Maitra A, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 2003; 4:437–450.
Goodwin GH, Johns EW . The isolation and purification of the high mobility group (HMG) nonhistone chromosomal proteins. Methods Cell Biol 1977; 16:257–267.
Lotze MT, Tracey KJ . High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol 2005; 5:331–342.
Muller S, Scaffidi P, Degryse B, et al. New EMBO members' review: the double life of HMGB1 chromatin protein: architectural factor and extracellular signal. EMBO J 2001; 20:4337–4340.
Dong XD, Ito N, Lotze MT, et al. High mobility group box I (HMGB1) release from tumor cells after treatment: implications for development of targeted chemoimmunotherapy. J Immunother 2007; 30:596–606.
Ellerman JE, Brown CK, de Vera M, et al. Masquerader: high mobility group box-1 and cancer. Clin Cancer Res 2007; 13:2836–2848.
Kang R, Chen R, Zhang Q, et al. HMGB1 in health and disease. Mol Aspects Med 2014; 40:1–116.
Andersson U, Tracey KJ . HMGB1 is a therapeutic target for sterile inflammation and infection. Annu Rev Immunol 2011; 29:139–162.
Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ . HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol 2010; 28:367–388.
Kang R, Zhang Q, Zeh HJ, 3rd, Lotze MT, Tang D . HMGB1 in cancer: good, bad, or both? Clin Cancer Res 2013; 19:4046–4057.
Taguchi A, Blood DC, del Toro G, et al. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature 2000; 405:354–360.
Bald T, Quast T, Landsberg J, et al. Ultraviolet-radiation-induced inflammation promotes angiotropism and metastasis in melanoma. Nature 2014; 507:109–113.
Kang R, Zhang Q, Hou W, et al. Intracellular Hmgb1 inhibits inflammatory nucleosome release and limits acute pancreatitis in mice. Gastroenterology 2014; 146:1097–1107.
Yadav D, Lowenfels AB . The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology 2013; 144:1252–1261.
Springer S, Wang Y, Dal Molin M, et al. A combination of molecular markers and clinical features improve the classification of pancreatic cysts. Gastroenterology 2015; 149:1501–1510.
Morris JPt, Wang SC, Hebrok M . KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat Rev Cancer 2010; 10:683–695.
Kopp JL, von Figura G, Mayes E, et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 2012; 22:737–750.
Crawford HC, Scoggins CR, Washington MK, Matrisian LM, Leach SD . Matrix metalloproteinase-7 is expressed by pancreatic cancer precursors and regulates acinar-to-ductal metaplasia in exocrine pancreas. J Clin Invest 2002; 109:1437–1444.
Bakhoum SF, Compton DA . Chromosomal instability and cancer: a complex relationship with therapeutic potential. J Clin Invest 2012; 122:1138–1143.
Lange SS, Mitchell DL, Vasquez KM . High mobility group protein B1 enhances DNA repair and chromatin modification after DNA damage. Proc Natl Acad Sci USA 2008; 105:10320–10325.
Celona B, Weiner A, Di Felice F, et al. Substantial histone reduction modulates genomewide nucleosomal occupancy and global transcriptional output. PLoS Biol 2011; 9:e1001086.
Bonner WM, Redon CE, Dickey JS, et al. GammaH2AX and cancer. Nat Rev Cancer 2008; 8:957–967.
Nishimura S . 8-Hydroxyguanine: From its discovery in 1983 to the present status. Proc Jpn Acad Ser B Phys Biol Sci 2006; 82:127–141.
Zambirinis CP, Pushalkar S, Saxena D, Miller G . Pancreatic cancer, inflammation, and microbiome. Cancer J 2014; 20:195–202.
Chen R, Kang R, Fan XG, Tang D . Release and activity of histone in diseases. Cell Death Dis 2014; 5:e1370.
Lesina M, Kurkowski MU, Ludes K, et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell 2011; 19:456–469.
Zhang Y, Yan W, Collins MA, et al. Interleukin-6 is required for pancreatic cancer progression by promoting MAPK signaling activation and oxidative stress resistance. Cancer Res 2013; 73:6359–6374.
Kang R, Loux T, Tang D, et al. The expression of the receptor for advanced glycation endproducts (RAGE) is permissive for early pancreatic neoplasia. Proc Natl Acad Sci USA 2012; 109:7031–7036.
Corcoran RB, Contino G, Deshpande V, et al. STAT3 plays a critical role in KRAS-induced pancreatic tumorigenesis. Cancer Res 2011; 71:5020–5029.
Fukuda A, Wang SC . Morris JP 4th, et al. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell 2011; 19:441–455.
Sirois CM, Jin T, Miller AL, et al. RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA. J Exp Med 2013; 210:2447–2463.
Xu J, Zhang X, Monestier M, Esmon NL, Esmon CT . Extracellular histones are mediators of death through TLR2 and TLR4 in mouse fatal liver injury. J Immunol 2011; 187:2626–2631.
Huang H, Evankovich J, Yan W, et al. Endogenous histones function as alarmins in sterile inflammatory liver injury through Toll-like receptor 9 in mice. Hepatology 2011; 54:999–1008.
Zambirinis CP, Levie E, Nguy S, et al. TLR9 ligation in pancreatic stellate cells promotes tumorigenesis. J Exp Med 2015; 212:2077–2094.
Mollica L, De Marchis F, Spitaleri A, et al. Glycyrrhizin binds to high-mobility group box 1 protein and inhibits its cytokine activities. Chem Biol 2007; 14:431–441.
Li XL, Zhou AG, Zhang L, Chen WJ . Antioxidant status and immune activity of glycyrrhizin in allergic rhinitis mice. Int J Mol Sci 2011; 12:905–916.
Collins MA, Bednar F, Zhang Y, et al. Oncogenic K-Ras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest 2012; 122:639–653.
Carriere C, Young AL, Gunn JR, Longnecker DS, Korc M . Acute pancreatitis accelerates initiation and progression to pancreatic cancer in mice expressing oncogenic Kras in the nestin cell lineage. PLoS One 2011; 6:e27725.
Hingorani SR, Wang L, Multani AS, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005; 7:469–483.
von Figura G, Fukuda A, Roy N, et al. The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat Cell Biol 2014; 16:255–267.
Rowell JP, Simpson KL, Stott K, Watson M, Thomas JO . HMGB1-facilitated p53 DNA binding occurs via HMG-Box/p53 transactivation domain interaction, regulated by the acidic tail. Structure 2012; 20:2014–2024.
Tang D, Kang R, Livesey KM, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol 2010; 190:881–892.
Tang D, Kang R, Livesey KM, et al. High-mobility group box 1 is essential for mitochondrial quality control. Cell Metab 2011; 13:701–711.
Bianchi ME . DAMPs, PAMPs and alarmins: all we need to know about danger. J Leukoc Biol 2007; 81:1–5.
Chung HW, Lim JB, Jang S, Lee KJ, Park KH, Song SY . Serum high mobility group box-1 is a powerful diagnostic and prognostic biomarker for pancreatic ductal adenocarcinoma. Cancer Sci 2012; 103:1714–1721.
Mefford HC, Trask BJ . The complex structure and dynamic evolution of human subtelomeres. Nat Rev Genet 2002; 3:91–102.
Polanska E, Dobsakova Z, Dvorackova M, Fajkus J, Stros M . HMGB1 gene knockout in mouse embryonic fibroblasts results in reduced telomerase activity and telomere dysfunction. Chromosoma 2012; 121:419–431.
Ke S, Zhou F, Yang H, et al. Downregulation of high mobility group box 1 modulates telomere homeostasis and increases the radiosensitivity of human breast cancer cells. Int J Oncol 2015; 46:1051–1058.
Quinlan JM, Colleypriest BJ, Farrant M, Tosh D . Epithelial metaplasia and the development of cancer. Biochim Biophys Acta 2007; 1776:10–21.
Morris JPt, Cano DA, Sekine S, Wang SC, Hebrok M . Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J Clin Invest 2010; 120:508–520.
Greer RL, Staley BK, Liou A, Hebrok M . Numb regulates acinar cell dedifferentiation and survival during pancreatic damage and acinar-to-ductal metaplasia. Gastroenterology 2013; 145:1088–1097 e1088.
Prevot PP, Simion A, Grimont A, et al. Role of the ductal transcription factors HNF6 and Sox9 in pancreatic acinar-to-ductal metaplasia. Gut 2012; 61:1723–1732.
Husain S, Thrower E . Molecular and cellular regulation of pancreatic acinar cell function. Curr Opin Gastroenterol 2009; 25:466–471.
Reichert M, Rustgi AK . Pancreatic ductal cells in development, regeneration, and neoplasia. J Clin Invest 2011; 121:4572–4578.
Cleveland MH, Sawyer JM, Afelik S, Jensen J, Leach SD . Exocrine ontogenies: on the development of pancreatic acinar, ductal and centroacinar cells. Semin Cell Dev Biol 2012; 23:711–719.
Rooman I, Real FX . Pancreatic ductal adenocarcinoma and acinar cells: a matter of differentiation and development? Gut 2012; 61:449–458.
Wei D, Wang L, Yan Y, et al. KLF4 is essential for induction of cellular identity change and acinar-to-ductal reprogramming during early pancreatic carcinogenesis. Cancer Cell 2016; 29:324–338.
Lynn FC, Smith SB, Wilson ME, Yang KY, Nekrep N, German MS . Sox9 coordinates a transcriptional network in pancreatic progenitor cells. Proc Natl Acad Sci USA 2007; 104:10500–10505.
Seymour PA, Freude KK, Tran MN, et al. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc Natl Acad Sci USA 2007; 104:1865–1870.
Shroff S, Rashid A, Wang H, Katz MH, Abbruzzese JL, Fleming JB . SOX9: a useful marker for pancreatic ductal lineage of pancreatic neoplasms. Hum Pathol 2014; 45:456–463.
Grimont A, Pinho AV, Cowley MJ, et al. SOX9 regulates ERBB signalling in pancreatic cancer development. Gut 2014; 64:1790–1799.
Chen NM, Singh G, Koenig A, et al. NFATc1 links EGFR signaling to induction of Sox9 transcription and acinar-ductal transdifferentiation in the pancreas. Gastroenterology 2015; 148:1024–1034.e9.
Chen R, Kang R, Fan XG, Tang D . Release and activity of histone in diseases. Cell Death Dis 2014; 5:e1370.
Fahmueller YN, Nagel D, Hoffmann RT, et al. Predictive and prognostic value of circulating nucleosomes and serum biomarkers in patients with metastasized colorectal cancer undergoing Selective Internal Radiation Therapy. BMC Cancer 2012; 12:5.
Roth C, Pantel K, Muller V, et al. Apoptosis-related deregulation of proteolytic activities and high serum levels of circulating nucleosomes and DNA in blood correlate with breast cancer progression. BMC Cancer 2011; 11:4.
Holdenrieder S, Von Pawel J, Nagel D, Stieber P . Long-term stability of circulating nucleosomes in serum. Anticancer Res 2010; 30:1613–1615.
Gebhardt C, Riehl A, Durchdewald M, et al. RAGE signaling sustains inflammation and promotes tumor development. J Exp Med 2008; 205:275–285.
Kang R, Chen R, Xie M, et al. The Receptor for Advanced Glycation End Products Activates the AIM2 Inflammasome in Acute Pancreatitis. J Immunol 2016; 196:4331–4337.
Kang R, Hou W, Zhang Q, et al. RAGE is essential for oncogenic KRAS-mediated hypoxic signaling in pancreatic cancer. Cell Death Dis 2014; 5:e1480.
Tian J, Avalos AM, Mao SY, et al. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol 2007; 8:487–496.
Chen R, Fu S, Fan XG, et al. Nuclear DAMP complex-mediated RAGE-dependent macrophage cell death. Biochem Biophys Res Commun 2015; 458:650–655.
Kang R, Tang D, Schapiro NE, et al. The HMGB1/RAGE inflammatory pathway promotes pancreatic tumor growth by regulating mitochondrial bioenergetics. Oncogene 2014; 33:567–577.
Zhu X, Messer JS, Wang Y, et al. Cytosolic HMGB1 controls the cellular autophagy/apoptosis checkpoint during inflammation. J Clin Invest 2015; 125:1098–1110.
Liliensiek B, Weigand MA, Bierhaus A, et al. Receptor for advanced glycation end products (RAGE) regulates sepsis but not the adaptive immune response. J Clin Invest 2004; 113:1641–1650.
Huang H, Tohme S, Al-Khafaji AB, et al. Damage-associated molecular pattern-activated neutrophil extracellular trap exacerbates sterile inflammatory liver injury. Hepatology 2015; 62:600–614.
Holdenrieder S, Stieber P, Bodenmuller H, et al. Nucleosomes in serum as a marker for cell death. Clin Chem Lab Med 2001; 39:596–605.
Ochi A, Nguyen AH, Bedrosian AS, et al. MyD88 inhibition amplifies dendritic cell capacity to promote pancreatic carcinogenesis via Th2 cells. J Exp Med 2012; 209:1671–1687.
Hruban RH, Adsay NV, Albores-Saavedra J, et al. Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol 2001; 25:579–586.
Acknowledgements
We thank Christine Heiner (Department of Surgery, University of Pittsburgh) for her critical reading of the manuscript. This work was supported by the National Institutes of Health (R01CA160417, R01GM115366, and R01CA181450), a Pancreatic Cancer Action Network-AACR Career Development Award (13-20-25-TANG), a Research Scholar Grant from the American Cancer Society (RSG-16-014-01-CDD), the National Natural Science Foundation of China (31671435), the National Natural Science Foundation of Guangdong (2016A030308), and the Central Finance Grant of China (16056001). This project partly utilized University of Pittsburgh Cancer Institute shared resources supported by award P30CA047904.
Author information
Authors and Affiliations
Corresponding authors
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
Generation of KCH mice. (PDF 120 kb)
Supplementary information, Figure S2
Histologic progression of pancreas shown by hematoxylin and eosin staining in KC, CH, KCH, and KCH+/− mice. (PDF 210 kb)
Supplementary information, Figure S3
Representative hematoxylin and eosin stains of pancreatic intraepithelial neoplasms (PanINs) (a), mucinous cystic neoplasms (MCNs) (b), intraductal papillary mucinous neoplasms (IPMNs) (c), and pancreatic ductal adenocarcinoma (PDAC) (d) in KCH mice at six weeks of age. (PDF 218 kb)
Supplementary information, Figure S4
Loss of HMGB1 promotes pancreatic function insufficiency during K-Ras-driven tumor development. (PDF 94 kb)
Supplementary information, Figure S5
KCH and KCH+/− mice exhibit significant acinar-to-ductal metaplasia (ADM). (PDF 182 kb)
Supplementary information, Figure S6
KCH and KCH+/− mice exhibit significant duct-like lesions. (PDF 146 kb)
Supplementary information, Figure S7
KCH and KCH+/− mice exhibit a significant stromal response. (PDF 121 kb)
Supplementary information, Figure S8
KCH and KCH+/− mice exhibit significant proliferation and invasion. (PDF 109 kb)
Supplementary information, Figure S9
Pancreata from KCH mice exhibit abnormal mRNA expression of genes involved in regulating the DNA damage response (A) and telomere maintenance (B) by RT2 Profiler™ PCR Array (n=3 mice/genotype). (PDF 96 kb)
Supplementary information, Figure S10
KCH mice exhibit abnormal expression of nucleosome receptors. (PDF 167 kb)
Supplementary information, Figure S11
KCHR mice (but not KCHT9 mice) exhibited decreased expression of SOX9, vimentin, Ki-67, and MMP7 in pancreas compared with KCH mice at 12 weeks of age (n=5 mice/genotype, ***p < 0.001, data are expressed as means ± s.e.m, unpaired t-test). (PDF 181 kb)
Supplementary information, Figure S12
(A) Loss of RAGE (but not TLR9) diminished recombinant nucleosomes-induced IL-6 release in isolated peritoneal macrophages and PDAC cells from indicated mice at 6 weeks of age (n=3, ***p < 0.001, data are expressed as means ± s.e.m, unpaired t-test versus KCH group). (PDF 56 kb)
Supplementary information, Table S1
Data on tumor metastasis and invasion in KC, KCH, and KCH+/− mice (PDF 23 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 Unported License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Kang, R., Xie, Y., Zhang, Q. et al. Intracellular HMGB1 as a novel tumor suppressor of pancreatic cancer. Cell Res 27, 916–932 (2017). https://doi.org/10.1038/cr.2017.51
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2017.51
Keywords
This article is cited by
-
The role of pyroptosis and gasdermin family in tumor progression and immune microenvironment
Experimental Hematology & Oncology (2023)
-
Inhibition of DAMP actions in the tumoral microenvironment using lactoferrin-glycyrrhizin conjugate for glioblastoma therapy
Biomaterials Research (2023)
-
PCDH1, a poor prognostic biomarker and potential target for pancreatic adenocarcinoma metastatic therapy
BMC Cancer (2023)
-
The multifunctional protein HMGB1: 50 years of discovery
Nature Reviews Immunology (2023)
-
Mechanisms of obesity- and diabetes mellitus-related pancreatic carcinogenesis: a comprehensive and systematic review
Signal Transduction and Targeted Therapy (2023)