Abstract
RGF1, a secreted peptide hormone, plays key roles in root meristem development in Arabidopsis. Previous studies indicated that a functional RGF1 needs to be sulfated at a tyrosine residue by a tyrosylprotein sulfotransferase and that RGF1 regulates the root meristem activity mainly via two downstream transcription factors, PLETHORA 1 (PLT1) and PLT2. How extracellular RGF1 is perceived by a plant cell, however, is unclear. Using genetic approaches, we discovered a clade of leucine-rich repeat receptor-like kinases, designated as RGF1 INSENSITIVE 1 (RGI1) to RGI5, serving as receptors of RGF1. Two independent rgi1 rgi2 rgi3 rgi4 rgi5 quintuple mutants display a consistent short primary root phenotype with a small size of meristem. An rgi1 rgi2 rgi3 rgi4 quadruple mutant shows a significantly reduced sensitivity to RGF1, and the quintuple mutant is completely insensitive to RGF1. The expression of PLT1 and PLT2 is almost undetectable in the quintuple mutant. Ectopic expression of PLT2 driven by an RGI2 promoter in the quintuple mutant greatly rescued its root meristem defects. One of the RGIs, RGI1, was subsequently analyzed biochemically in detail. In vitro dot blotting and pull-down analyses indicated that RGI1 can physically interact with RGF1. Exogenous application of RGF1 can quickly and simultaneously induce the phosphorylation and ubiquitination of RGI1, indicating that RGI1 can perceive and transduce the RGF1 peptide signal. Yet, the activated RGI1 is likely turned over rapidly. These results demonstrate that RGIs, acting as the receptors of RGF1, play essential roles in RGF1-PLT-mediated root meristem development in Arabidopsis thaliana.
Similar content being viewed by others
Introduction
Cell-cell communication is crucial for normal plant growth and development. Higher plants, like other multicellular organisms, are often developed from one single cell, a fertilized egg, which gradually develops into a sophisticated and well-organized individual with all necessary tissues and organs. These developmental processes require constant intercellular communications. Plant cells can synthesize various peptide hormones as signaling molecules. These peptides can be delivered to apoplast mainly for short-distance communications in a non-cell autonomous manner1. There are two major classes of secreted peptide signals, including small peptides with 5-20 amino acids and cysteine-rich peptides with ∼50 amino acids2. The peptides of the former class are often proteolytically processed from their larger precursor proteins, pre-proproteins, and usually require posttranslational modifications such as glycosylation, proline hydroxylation, and tyrosine sulfation1. A pre-proprotein often contains a hydrophobic signal sequence at its N-terminus, leading the precursor to the secretory system. The maturation of a peptide hormone typically requires several steps. A pre-proprotein is cleaved by a peptidase in the endoplasmic reticulum to remove the N-terminal peptide sequence and subsequently forms a proprotein, which is then modified and packaged into membrane-bound secretory vesicles3. Specific endoproteases can then cleave the proprotein to form an active peptide hormone, which is secreted into apoplast and perceived by cell surface receptors such as leucine-rich repeat (LRR) receptor-like kinases (RLKs; LRR-RLKs)4,5,6.
In the past two decades, an increasing amount of peptide hormones have been identified in plants. They play significant roles in regulating many different aspects of plant growth and development, as well as defense responses against biotic and abiotic stresses1. Among them, CLAVATA3 (CLV3)/endosperm surrounding region (ESR) (CLE) and CLE-like (CLEL), two related but clearly distinct groups of peptide hormones, have received more attention than any other peptide hormones. There are 32 CLE genes in the Arabidopsis genome7. Promoter activity analyses indicated that almost all tissues show the expression of some of the CLE genes, suggesting their broad biological functions8. All CLE peptides are cleaved from larger CLE precursor proteins. A typical CLE precursor protein contains two major domains, an N-terminal signal peptide sequence for directing the protein to the secretory pathway and a C-terminal CLE domain containing about 13-14 amino acids7,9,10. Biological functions of several CLE peptides have been revealed. For example, CLV3 and CLE40 are responsible for shoot and root apical meristem maintenance, respectively9,11,12. CLV3 is perceived by three different receptors or putative receptors, CLAVATA1 (CLV1), CLAVATA 2 (CLV2)-CORYNE (CRN), and RPK2/TOAD2, to limit stem cell size by restricting the expression of a homeodomain transcription factor, WUSCHEL (WUS), via a negative feedback loop13,14,15,16,17,18. On the other hand, CLE40 is perceived by its putative receptor complex containing two different RLKs, ARABIDOPSIS CRIKLY4 (ACR4) and CLV1, to inhibit the expression of a root version of homeodomain transcription factor, WOX519,20. TRACHEARY ELEMENT DIFFERENTIATION INHIBITORY FACTOR (TDIF)/CLE41/CLE44 peptide regulates vascular development in Arabidopsis. TDIF peptide derived from CLE41 or CLE44 is perceived by PHLOEM INTERCALATED WITH XYLEM (PXY), an LRR-RLK21,22,23,24.
A group of Arabidopsis CLE-related peptides were identified via different searching strategies. These peptides are expressed in the root meristem region and are responsible for root meristem maintenance. This family of peptides was initially designated as ROOT MERISTEM GROWTH FACTORS (RGFs)25. The same family of peptides was also named CLEL because of the CLEL motifs at their C-termini26, or GOLVEN (GLV) as transgenic plants overexpressing RGF precursor genes showed wavy root phenotypes on slanted plates, suggesting their roles in regulating plant gravitropic responses27. RGF/CLEL/GLV peptide family contains 11 members28. For simplicity, we use RGFs hereafter. Like CLE precursor proteins, RGF precursors also contain two major domains, an N-terminal signal peptide sequence and a C-terminal RGF peptide domain26. The active peptides contain about 13 amino acids with an Asp-Tyr signature critical for tyrosine sulfation, which is required for the activity of RGFs25. There is only one gene encoding tyrosylprotein sulfotransferase (TPST) in Arabidopsis genome29. tpst-1 mutant shows a short root phenotype with significantly reduced meristematic activity25,30. The reduced meristem size of tpst-1 mutant can be partially rescued by exogenous application of synthetic sulfated RGF125. In fact, in the presence of sulfated RGF1 together with two other sulfated peptides, PSK and PSY1, the root growth defects of tpst-1 mutant can be fully recovered25. It was reported that auxin can induce the expression of TPST30. RGF1 can be activated by TPST and the activated RGF1 can stabilize two AP1 domain-containing transcription factors, PLATHOLA 1 (PLT1) and PLT2, or induce the expression of their corresponding genes25. A main gap in this signaling cascade is unidentified receptor/receptors of RGF1.
RLKs play key roles in regulating plant growth and development as well as adaptations to numerous biotic and abiotic stresses. There are ∼223 LRR-RLKs in Arabidopsis31. Only about 60 LRR-RLKs have been functionally reported up-to-date32. A vast majority of LRR-RLKs still need to be functionally characterized. Our laboratory is interested in revealing the biological functions of these undefined LRR-RLKs. Our previous genetic analyses indicated that BAK1 and its paralogs are involved in regulating brassinosteroid (BR)-independent root growth and development, as serk1 serk2 bak1 and serk1 bak1 bkk1 triple null mutants show intensively shorter roots compared with BR-deficient or BR signaling-impaired mutants33,34. We proposed that BAK1 and its paralogs may serve as a co-receptor and pair with another LRR-RLK to regulate root growth and development, similar to its role in BRI1- or FLS2-mediated signaling pathway35,36,37,38,39,40,41,42,43. Using a yeast two-hybrid approach, we identified dozens of LRR-RLKs which can physically interact with BAK1. Among them are 5 closely related LRR-RLKs in the LRR XI subfamily (data not shown). We therefore used a reverse genetic approach to test whether they regulate root development. We generated two sets of independent quintuple mutants. Phenotypic analyses revealed that they exhibited an extremely short root phenotype, with only a few meristematic cortex cells. Unlike the tpst-1 mutant, the quintuple mutant is completely insensitive to exogenously applied synthetic sulfated RGF1. These five LRR-RLKs were then named RGF1 INSENSITIVEs (RGIs). Detailed analyses using one of the RGIs, RGI1, indicated that RGF1 can physically interact with the extracellular domain of RGI1. Treatment of transgenic plants harboring 35S::RGI1-FLAG with synthetic sulfated RGF1 for 30 min can significantly induce the phosphorylation and ubiquitination of RGI1-FLAG. Our data clearly indicate that RGIs play redundant roles in controlling root meristem development. At least one of the RGIs, RGI1, has been demonstrated, both genetically and biochemically, as the receptor of RGF1.
Results
Knockout of five LRR-RLKs from the LRR XI subfamily in a single plant resulted in an extremely short root phenotype
Our previous analysis indicated that BAK1 and its paralogs are involved in root growth and development, which is independent of the BR signaling pathway34. BAK1 was identified as a common co-receptor in a number of RLK-mediated signaling pathways35,36,37,38,39,40,41,42,43. We therefore hypothesized that BAK1 may pair with another ligand-interacting LRR-RLK to regulate root development34. To identify this putative LRR-RLK which can physically interact with BAK1, we carried out yeast two-hybrid analysis to test the interaction of BAK1 with all other 161 LRR-RLKs, whose cDNAs were cloned previously44. Using BAK1 as bait, we totally identified 83 LRR-RLKs which can interact with BAK1 in an mbSUS yeast two-hybrid system45. Interestingly, a clade of 5 LRR-RLKs in the LRR XI subfamily, At3G24240, At5G48940, At4G26540, At5G56040, and At1G34110, all show some degree of interaction with BAK1 (data not shown). Therefore, we decided to study the biological functions of these 5 LRR-RLKs, which were later named RGI1 to RGI5 (Figure 1A) because our genetic analyses indicated that the quintuple mutant of these 5 LRR-RLKs is completely insensitive to RGF1. Hereafter, we use RGI1 to RGI5 to refer to these 5 LRR-RLKs.
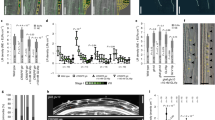
Quintuple mutants of five closely related LRR-RLKs, RGI1 to RGI5, show extremely short root and small meristem. (A) A phylogenetic tree of RGI1 to RGI5 based on kinase domain amino acid sequences of the five LRR-RLKs. SOBIR1 was used as an outgroup control. (B) Two independent sets of T-DNA insertion lines were used to generate two independent quintuple mutants. The allele names marked in red were used to generate the first quintuple mutant and the allele names marked in blue were used to generate the second independent quintuple mutant. (C) Two independent quintuple mutants and their crossed F1 heterozygous seedlings all show extremely short root phenotypes. To simplify, the first set of quintuple mutant was also named rgi1,2,3,4,5. The seedlings were photographed 5 days after germination. Scale bar, 5 mm. (D) Measurements of the roots of Col-0 and two independent quintuple mutants shown in C. Data shown are mean ± SD (n > 40). Student's t-tests were performed based on their differences with the wild-type control (***P < 0.001). (E) PI-stained root tips of Col-0 and two independent quintuple mutants. Scale bar, 50 μm. (F) Numbers of meristematic cortex cells of Col-0 and two independent quintuple mutants. Data shown are mean ± SD (n > 15). Student's t-tests were performed based on their differences with the wild-type control (***P < 0.001).
Gene structure analyses showed that all five RGI genomic sequences contain only one intron close to their 3′ stop codons. Their coding regions encode proteins with 1 141, 1 135, 1 091, 1 090, and 1 072 amino acids. The variable amino acid sequences among 5 RGIs mainly locate at their N-terminal signal peptides and their C-terminal tails. These five RGIs are highly conserved at the amino acid level. For example, RGI1 share 63%, 49%, 49%, and 47% amino acid identity and 77%, 68%, 68%, and 64% amino acid similarity with RGI2, RGI3, RGI4, and RGI5, respectively. Domain identification analyses using the SMART software in combination with manual counting revealed that all five RGIs contain 25 LRRs within their extracellular domains.
To understand biological roles of these RGIs, we first checked their expression patterns and found that all five genes are expressed in roots, four of which can be detected in the root tips, with the exception of RGI3 (Supplementary information, Figure S1A)32. We isolated two independent sets of T-DNA insertion lines for each of these 5 RGIs (Figure 1B). RT-PCR analyses confirmed that all of them cannot express full-length cDNAs (Supplementary information, Figure S1B). None of the single mutant showed any significant defective phenotypes compared with the wild-type plant. We therefore generated two independent series of double, triple, quadruple, and quintuple mutants. Morphological analyses indicated that the defective phenotypes are mainly observed in the root but not significantly in the aerial parts (Supplementary information, Figure S1C-S1E), with quintuple mutant showing the most severe root developmental defects (Figure 1C and 1D), which is consistent with their expression profiles (Supplementary information, Figure S1A)32. Two independent quintuple mutants, rgi1-1 rgi2-1 rgi3-1 rgi4-1 rgi5-1 and rgi1-2 rgi2-2 rgi3-2 rgi4-2 rgi5-2, showed almost identical short root phenotypes, which are only about 20% the length of wild-type plant, Col-0 (Figure 1C and 1D). Interestingly, F1 plants generated by crossing these two quintuple mutants also showed a similar short root phenotype, further confirming that the root defects were caused by the disruption of the 5 RGIs (Figure 1C and 1D). Confocal microscopic analyses of propidium iodide (PI)-stained root tips also showed that the meristem size is greatly reduced in the quintuple mutants compared with wild-type seedlings (Figure 1E and 1F). Five days after germination, wild-type seedlings contain about 25 meristematic cortex cells but the number for quintuple mutants is only 4-5 (Figure 1F). Because two quintuple mutants showed almost identical phenotypes, all our later experiments were carried out by using the first quintuple mutant, rgi1-1 rgi2-1 rgi3-1 rgi4-1 rgi5-1, or simplified as rgi1,2,3,4,5. Next we performed the complementation experiments in the quintuple mutant. Expression of RGI2 (driven by its own promoter), one of the most abundantly expressed RGIs in the root meristem zone, can greatly reverse the short root and small meristem phenotype (Figure 2A-2D and Supplementary information, Figure S2). These genetic data suggest the critical roles of 5 RGIs in regulating root meristem maintenance.
The short root phenotype of the quintuple mutant can be rescued via the expression of RGI2 driven by its native promoter. (A) The root phenotypes of Col-0, rgi1,2,3,4,5 quintuple mutant, and two independent transgenic lines of the quintuple mutant with expression of RGI2 grown on a 1/2 MS medium. The seedlings were photographed 6 days after germination. Scale bar, 5 mm. (B) Measurement of the roots of Col-0, quintuple mutant, and two independent lines shown in A. Data shown are mean ± SD (n > 30). Student's t-tests were performed based on the differences of each of the two independent lines with the quintuple mutant (***P < 0.001). (C) PI-stained root tips of Col-0, quintuple mutant, and two independent lines shown in A. Roots from seedlings 4 days after germination were used for the staining. White arrow heads mark the junction of meristem and elongation zones. Scale bar, 50 μm. (D) Meristematic cortex cell numbers of Col-0, quintuple mutant, and two independent lines shown in A. Data shown are mean ± SD (n > 15). Student's t-tests were performed based on the differences of each of the two transgenic lines with the quintuple mutant (***P < 0.001).
Expression levels of PLT1 and PLT2 were greatly downregulated in the quintuple mutant
There are several parallel signaling pathways involved in root stem cell niche maintenance, such as SHR-SCR46,47,48,49,50, CLE40-ACR4/CLV1-WOX519,20, and RGF1-PLT pathways25,30,51,52. To examine which pathway was mainly affected in the quintuple mutant, we crossed pSHR::NLS-YFP, pSCR::NLS-YFP, pWOX5::NLS-YFP, pPLT1::CFP, pPLT2::CFP, pPLT1::PLT1-YFP, and pPLT2::PLT2-YFP transgenic plants with Col-0 and the quintuple mutant. Using the same individual transgenic line to cross with wild-type plant and the quintuple mutant, positional effect can be controlled. The expression levels of the marker genes in Col-0 were then compared with those in the quintuple mutant.
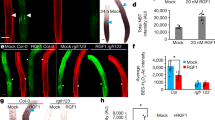
The expression levels and patterns of both SHR and SCR are not significantly altered in the quintuple mutant compared with the wild-type plant (Supplementary information, Figure S3A and S3B), which is consistent with root cross section results, showing that there are no obvious cell differentiation defects in the quintuple mutant (Supplementary information, Figure S1C). The expression domain of WOX5 in the quintuple mutant, however, is greatly expanded (Supplementary information, Figure S3C), suggesting that there are more quiescent center (QC) cells in the quintuple mutant than wild-type plant. In addition, the promoter activities of PLT1 and PLT2 are significantly downregulated in the quintuple mutant (Figure 3A-3D). Protein expression of PLT1-YFP and PLT2-YFP is almost undetectable in the quintuple mutant (Figure 3E-3H). Further genetic assay indicated that WOX5 expression domain expansion is not the cause of the short root phenotype, as rgi1,2,3,4,5 wox5 sextuple mutant showed more severe but not suppressed phenotype (Supplementary information, Figure S3D). On the other hand, ectopic expression of PLT2 driven by the RGI2 promoter can completely reverse the meristem size defect in the rgi1,2,3,4 quadruple mutant and can greatly suppress that in the quintuple mutant (Figure 4A-4D). These results demonstrate that the RGF1-PLT pathway is blocked in the quintuple mutant, which is most likely the main cause of the largely reduced meristem size phenotype.
The expression levels of PLT1 and PLT2 are significantly downregulated in the quintuple mutant compared with the wild-type plant. (A-H) Representative roots showing the expression of pPLT1::CFP (A, B), pPLT2::CFP (C, D), pPLT1::PLT1-YFP (E, F) and pPLT2::PLT2-YFP (G, H) in Col-0 and the quintuple mutant. Roots are from seedlings 4 days after germination. Scale bar, 50 μm.
Ectopic expression of PLT2 driven by the promoter of RGI2 completely rescues the meristem size of rgi1,2,3,4 and partially rescues the meristem size of rgi1,2,3,4,5. (A, B) PI-stained root tips of Col-0, quadruple mutant, and two independent transgenic lines and their measurements. White arrow heads indicate the junction between meristem and elongation zones. The roots are from seedlings 5 days after germination. Scale bar, 50 μm. Student's t-tests were performed based on the differences between each of the two lines of transgenic seedlings and the quadruple mutant (***P < 0.001, n > 15). (C, D) PI-stained root tips of Col-0, quintuple mutant, and two independent transgenic lines and their measurements. White arrow heads indicate the junction between meristem and elongation zones. The roots are from seedlings 4 days after germination. Scale bar, 50 μm. Student's t-tests were performed based on the differences between each of the two lines of transgenic seedlings and the quintuple mutant (***P < 0.001, n > 15).
To further confirm that RGIs are the key components in the RGF1-PLT pathway, we compared the root phenotypes of rgi1,2,3,4,5 with tpst-1 and plt1-4 plt2-2. All these mutants showed very similar short root phenotype (Supplementary information, Figure S4A and S4B). Microscopic analyses revealed that all these mutants exhibited significantly reduced meristem size (Supplementary information, Figure S4C and S4D). We also generated rgi1,2,3,4,5 tpst-1 sextuple mutant and found that it did not show more severe root defective phenotype (Supplementary information, Figure S4E and S4F). These genetic data support that RGIs, like TPST, mainly regulate the RGF1-PLT pathway.
The quintuple mutant is insensitive to exogenously applied synthetic sulfated RGF1
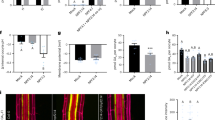
The major missing component in the RGF1-PLT pathway is the receptor of the peptide RGF1. Based on the aforementioned genetic results and the biochemical property of RGIs, we hypothesized that RGIs are the receptors of RGF1. To test this hypothesis, we treated both Col-0 and the rgi1,2,3,4 quadruple mutant with synthesized sulfated RGF1. Consistent with previous reports, wild-type seedlings showed altered gravitropism response, reduced lateral root formation, and dramatically increased number of meristem cortex cells after the treatment25,26,27,53. The quadruple mutant, on the other hand, showed no gravitropism alteration, unchanged lateral root formation, and subtly increased number of meristem cortex cells (Figure 5A-5D). When rgi1,2,3,4,5 was treated with RGF1, the number of meristem cortex cells showed no alteration, suggesting that the quintuple mutant is completely insensitive to RGF1 (Figure 5E and 5F). Consistently, qRT-PCR analysis indicated that the expression levels of PLT1 and PLT2 can be upregulated in wild-type plant and tpst-1 mutant, but not in the quintuple mutant, after the RGF1 treatment (Supplementary information, Figure S5A and S5B). In other words, the entire RGF1-PLT signaling pathway is blocked in the quintuple mutant.
rgi1,2,3,4 quadruple mutant shows reduced sensitivity to RGF1, and rgi1,2,3,4,5 quintuple mutant is completely insensitive to RGF1. (A) Phenotypes of Col-0 and the quadruple mutant growing on 1/2 MS medium without (up panel) and with 200 nM RGF1 (bottom panel). Scale bar, 1 cm. (B) Measurements of lateral root numbers of Col-0 and the quadruple mutant in the presence or absence of 1 μM RGF1. Student's t-tests were performed based on the differences between RGF1-treated and -untreated seedlings (***P < 0.001, ns represents no significant difference, n> 40). The measurements were carried out 10 days after germination. (C, D) Response of meristem size of Col-0 and the quadruple mutant to the treatment of 100 nM RGF1. The roots are from seedlings 5 days after germination. Data shown in D represent the measurements from C. Scale bar in C, 50 μm. Student's t-tests were performed based on the differences between RGF1-treated and -untreated seedlings (***P < 0.001, **P < 0.01, n> 20). (E, F) Response of meristem size of Col-0 and the quintuple mutant to the treatment of 100 nM RGF1. The roots are from seedlings 5 days after germination. Data shown in F represent the measurements from E. Scale bar in E, 50 μm. Student's t-tests were performed based on the differences between RGF1-treated and -untreated seedlings (***P < 0.001, ns represents no significant difference, n > 20).
RGF1 interacts with RGI1 and induces its phosphorylation and ubiquitination
To further confirm that RGIs are the receptors of RGF1, we first demonstrated that RGI1 is indeed a plasma membrane-localized RLK using 35S::RGI1-GFP transgenic plants (Supplementary information, Figure S6A). Lacking an antibody against RGF1, we synthesized a sulfated FLAG-RGF1 and confirmed its biological function via the meristem stimulation activity assay (Supplementary information, Figure S6B and S6C). We subsequently carried out dot blotting assays by immobilizing MBP and MBP-RGI1ED (extracellular domain), purified from E. coli, on a piece of nitrocellulose membrane, and used FLAG-RGF1 as a probe. Our result showed that MBP-RGI1ED, but not MBP, can associate with FLAG-RGF1 (Figure 6A). Pull-down analysis further confirmed the interaction between MBP-RGI1ED and FLAG-RGF1 (Supplementary information, Figure S6D).
RGI1 interacts with RGF1, and RGF1 induces the phosphorylation and poly-ubiquitination of RGI1. (A) Dot blotting assay showing that MBP-RGI1ED, but not MBP, can interact with FLAG-RGF1. Top panel shows Commassie blue-stained 100 ng column-purified MBP and MBP-RGI1ED after SDS-PAGE. Bottom panel shows that immobilized 100 ng MBP-RGI1ED but not MBP can interact with the FLAG-RGF1 probe. An anti-FLAG antibody was used for the immunoblotting assay. (B) E. coli-expressed MBP-RGI1CD showing strong autophosphorylation signal. Mutation of K814E but not K815E abolished its autophosphorylation. (C) A transgenic line harboring 35S::RGI1-FLAG shows induced phosphorylation of RGI1-FLAG after the treatment of 20 μM RGF1 for 30 min in a 1/2 MS medium liquid culture. (D) The 35S::RGI1-FLAG transgenic line was treated with or without 20 μM RGF1 for 30 min. RGI1-FLAG was immunoprecipitated and treated with or without CIP. The proteins were then subjected to western blot analysis using anti-FLAG antibody. (E) RGF1 treatment can induce the poly-ubiquitination of RGI1. Top panel shows the immunoblotting result using an anti-ubiquitin antibody. Bottom panel is the immunoblotting result using an anti-FLAG antibody. (F) RGF1 treatment can significantly reduce the protein level of RGI1. Three-week-old Arabidopsis rosette leaves were treated with or without 20 μM RGF1 for 30 min before total protein was extracted and analyzed by immunoblotting.
RGI1CD (cytoplasmic domain (CD)) contains all 11 subdomains that are conserved for serine/threonine kinases54. To test whether RGI1 has kinase activity, we cloned the intracellular cytoplasmic domain of RGI1 and fused it with MBP. Western blotting analysis using an anti-phosphothreonine antibody showed that MBP-RGI1CD has strong autophosphorylation activity. There are two lysine residues in the kinase subdomain II, K814 and K815. Previous studies indicated that mutation of a conserved lysine residue in this subdomain of several LRR-RLKs, such as BAK1 and BRI1, can completely abolish their kinase autophosphorylation activities35,54. We therefore made two independent mutations, K814E and K815E. MBP-RGI1CD K815E showed normal autophosphorylation, similar to that of MBP-RGI1CD. However, MBP-RGI1CD K814E completely lost autophosphorylation activity (Figure 6B), suggesting that K814 is the conserved residue critical for the kinase activity of RGI1.
To examine whether RGI1 can transduce the RGF1 signal from its extracellular domain to intracellular domain, we generated transgenic plants overexpressing RGI1-FLAG under the control of a constitutive CaMV 35S promoter. Phosphorylation level of RGI1-FLAG was drastically increased after treatment with RGF1 for 30 min in liquid culture, as determined by the western blotting assay using anti-pThr antibody (Figure 6C). When an anti-FLAG antibody was used for analyzing loading control, two bands appeared after RGF1 treatment. To test whether the top band is a phosphorylated form of RGI1, we treated the sample with calf intestine phosphatase (CIP), a phosphatase with broad-spectrum activity. Unexpectedly, the top band moved only slightly down but not completely after CIP treatment, suggesting that there are other modifications besides phosphorylation (Figure 6D). We therefore tested whether the top band also contains ubiquitinated RGI1 by using a specific anti-ubiquitin antibody. Interestingly, upon RGF1 treatment, RGI1 showed dramatically increased poly-ubiquitination (Figure 6E). In addition, western blotting analysis indicated that RGF1 treatment can lead to the degradation of RGI1 (Figure 6F). Taken together, these results clearly demonstrate that RGI1 is a bona fide receptor of RGF1, which not only can physically interact with RGF1 but also can transduce an extracellular signal to intracellular biochemical responses including phosphorylation and ubiquitination.
Discussion
Our genetic and biochemical analyses revealed that RGIs are key regulators in the RGF1-PLT signaling pathway, most likely as receptors of RGF1, to control root meristem development. First, two sets of independent quintuple homozygous mutants and the F1 plants generated via crossing these two mutants showed almost identical root defective phenotypes (Figure 1C-1F). The root development defects in the quintuple mutant can be reversed by re-introduction of RGI2 (Figure 2A-2D). These data demonstrate that the root defective phenotype is caused by the disruption of 5 RGIs and unlikely by other unknown mutations. Second, our genetic analyses also corroborated that it is the block of the RGF1-PLT but not the SHR-SCR or CLE40-ACR4 pathway which caused the root defective phenotype observed in the quintuple mutants (Supplementary information, Figures S1C and S3A-S3D). In the quintuple mutant, the expression of PLT1 and PLT2 is almost undetectable (Figure 3A-3H). Ectopic expression of PLT2 driven by an RGI2 promoter in the quintuple mutant can greatly reverse the defective phenotypes of the quintuple mutant (Figure 4C and 4D), suggesting that RGIs act upstream of PLTs. In addition, null mutations in other components within the RGF1-PLT signaling pathway gave rise to root defective phenotype very similar to that of the quintuple mutant (Supplementary information, Figure S4A-S4D). Third, exogenous application of synthetic sulfated RGF1 can induce multiple changes in wild-type plants, including upregulated expression of PLT1 and PLT2 (Supplementary information, Figure S5A and S5B), increased meristematic cortex cell number (Figure 5C-5F), reduced lateral root numbers and altered gravitropism response (Figure 5A and 5B); however, all these RGF1-induced responses were barely observed in the quadruple or quintuple mutant (Figure 5A-5F, Supplementary information, Figure S5A and S5B). These observations all demonstrate that RGIs are essential components in the RGF1-PLT signaling pathway. The protein properties of RGIs as LRR-RLKs suggest that they are likely involved in the perception of RGF1. Indeed, dot blotting and in vitro pull-down assays revealed that synthetic sulfated FLAG-RGF1 can interact with MBP-RGI1ED but not MBP (Figure 6A and Supplementary information, Figure S6D). Finally, our results showed that RGF1 can quickly induce the phosphorylation and ubiquitination of RGI1 (Figure 6C-6E), suggesting that RGI1 not only can perceive the RGF1 signal in the extracellular space but also can transduce the signal into the intracellular compartment via protein phosphorylation. Therefore, we believe that RGIs, at least RGI1, qualify as the receptors of RGF1. This conclusion is consistent with recent independent studies from two other research groups55,56. Matsubayashi's group used a photoaffinity approach and identified three RGF1 receptors in Arabidopsis55, which are identical to our RGI1 to RGI3. On the other hand, Chai's group identified the same 5 RGF1 receptors as we show here based on the presence of conserved RXR and RXGG signature motifs in the extracellular domains of these receptors56. The RXR and RXGG motifs can interact with the C-terminal asparagine residue and the sulfate group of RGF1, respectively. In addition, they solved the structure of the RGF1-At4G26540 complex (equivalent to our RGI3) at a resolution of 2.6 Å56.
Previous studies indicated that the perception of BR and flg22 by their receptors BRI1 and FLS2 often results in the recruitment of a co-receptor, such as BAK135,36,39,40,43. The formation of a receptor-ligand-co-receptor complex leads to a fully activated BR or flg22 downstream signaling cascade. Similarly, we hypothesize that the association of sulfated RGF1 with the extracellular domain of RGIs may induce the recruitment of BAK1 and its paralogs as co-receptors to fully activate the kinase activity of RGIs (Figure 7). The activated RGIs can then transduce the signal to downstream components and regulate the accumulation of two critical transcription factors, PLT1 and PLT2, thus controlling root meristem development. The notion that BAK1 and its paralogs may be the co-receptors of RGIs is supported by a few observations. First, serk1 bak1 bkk1 or serk1 serk2 bak1 triple mutants show short root phenotype which is independent of the BR signaling pathway33,34. Second, members of RGIs were found to interact with BAK1 in a yeast two-hybrid analysis (data not shown), which was the primary motivation prompted us to study the functions of the entire clade of RGIs. Third, all BAK1 and its paralogs were found to be expressed in the roots32. Finally, most of the characterized ligand-binding LRR-RLKs use BAK1 and its paralogs as their co-receptors. Consistently, biochemical analysis by Chai's group showed that these five LRR-RLKs can form heterodimers with SERK family members including BAK1 upon ligand induction and genetic studies further confirmed that these SERKs serve as co-receptors in this signaling pathway56.
A hypothetical model showing the role of RGI1 in regulating RGF1-PLT-mediated root development. Auxin upregulates the transcription of TPST. TPST can sulfate and activate RGF1. The activated RGF1 can be secreted from the cell by exocytosis and perceived by RGI1. Upon the binding, RGI1 may recruit a co-receptor, likely BAK1 or its homologs. The intracellular kinases of both RGI1 and its putative co-receptor can be activated via protein phosphorylation. The activated RGI1 can further activate its yet unidentified regulators and ultimately induce the expression of two AP1-domain transcription factors and control root meristem development. At the same time, activated RGI1 is poly-ubiquitinated and degraded via the 26S proteasome pathway.
It is interesting that upon the treatment of synthesized RGF1, both phosphorylation and poly-ubiquitination of RGI1 were quickly induced. Although about 10 pairs of ligand-RLK have been revealed, only a few receptors show ligand-induced phosphorylation. Many RLK receptors were found to be kinase-dead receptors. The best characterized RLK receptor and co-receptor are probably BRI1 and BAK1, the receptor and co-receptor of BRs, respectively35,36,57. Phosphorylation of BRI1 and BAK1 can be induced rapidly37,58. In fact, the activation of BRI1 and BAK1 is via a sequential phosphorylation mechanism37. To our knowledge, RGI1 represents probably the first LRR-RLK whose phosphorylation and poly-ubiquitination can be rapidly induced simultaneously. Poly-ubiquitination of a protein often suggests that the protein is targeted to the 26S proteasome degradation pathway. This result suggests when RGI1 is activated, it needs to be degraded. Otherwise it may be detrimental to normal root growth and development.
Phenotypic similarity of tpst-1, rgi1,2,3,4,5, and plt1-4 plt2-2 supports that TPST, RGF1, RGIs, and PLT are in the same signaling pathway. Our results showed that the phenotype of tpst-1 is slightly less severe than the quintuple mutant. This is probably because some unsulfated CLEL peptides, such as CLEL6, may still possess some degree of activity as suggested in a previous report26. The tpst-1 and plt1-4 plt2-2 mutants showed root defects similar to the quintuple mutant. In addition, the expression of PLT1 and PLT2 is almost undetectable in the quintuple mutant. These observations suggest that RGIs are the main components controlling the expression of PLT1 and PLT2.
In summary, identification of the receptors of the RGF peptides opens up a new avenue for future investigation. Numerous questions regarding the functions of RGF/CLEL/GLV peptides will be gradually elucidated. The studies reported here only focus on the roles of RGFs in regulating root meristem development. RGFs are also involved in a variety of other biological functions which have not been explored in detail. For example, overexpression of RGF/CLEL/GLV or exogenous application of RGF peptides often results in defective root gravitropic phenotypes. But the underlying molecular mechanisms are not understood. It is difficult to link peptide hormones to auxin transporters or auxin signaling. With the discovery of the RGF receptors, it opens up opportunities for these analyses in the near future. In addition, it will also be very interesting to investigate the entire signaling pathway from the perception of RGFs by RGI receptors to PLT expression. For example, future investigation may focus on identifying the substrates of RGIs, F-box proteins mediating poly-ubiquitination of RGIs, and the transcription factors directly regulating the expression of PLT1 and PLT2.
Materials and Methods
Plant materials and growth conditions
The Arabidopsis Col-0 or WS2 was used as a wild-type control in the experiment. The T-DNA insertion lines, including rgi1-1(SALK_020659), rgi2-1 (SALK_039309), rgi3-1(SALK_053167C), rgi4-1(SALK_052069C), rgi5-1(SALK_058918C), rgi1-2(SALK_040393C), rgi2-2(SALK_097109), rgi3-2(SAIL_1220_B03), rgi4-2(SALK_146219), rgi5-2(SALK_014726), and wox5(SALK_038262C) were purchased from the Arabidopsis Biological Resource Center (ABRC). Homozygous T-DNA insertion lines were isolated and confirmed by PCR and used for genetic crossing. The seeds of tpst-1 (also named aqc1-2), plt1-4 plt2-2, and transgenic plants harboring pPLT1::CFP, pPLT2::CFP, pPLT1::PLT1-YFP, and pPLT2::PLT2-YFP were provided by Professor Chuanyou Li (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China). The promoters of RGI1, RGI2, RGI3, RGI4, and RGI5 were amplified by PCR and cloned into a pBIB-BASTA-GUS-GWR vector. The obtained transgenic plants were used for GUS staining as described previously32. The promoter sequences of SHR, SCR, and WOX5 were amplified by PCR and cloned into a pBIB-BASTA-NLS-YFP-GWR vector and the obtained transgenic plants were used for confocal imaging. All transgenic lines were initially generated in Col-0 background and the single insertion line was selected, which was used for crossing with the rgi1-1 rgi2-1 rgi3-1 rgi4-1 rgi5-1 quintuple mutant to avoid position effect. A 5.6 kb fragment including 1.6 kb promoter of RGI2 genomic DNA was cloned into a pBIB-BASTA-FLAG-GWR vector for complementation. The same promoter was used to drive the expression of PLT2 CDS in rgi1-1 rgi2-1 rgi3-1 rgi4-1 and rgi1-1 rgi2-1 rgi3-1 rgi4-1 rgi5-1 for testing whether it can rescue the root meristem defects. The CDS of RGI1 was amplified and cloned into PBIB-BASTA-35S-FLAG-GWR and PBIB-BASTA-35S-GFP-GWR vectors, respectively, for overexpressing RGI1 in transgenic plants. All transgenic plants were generated through a floral dip method59. All primers used for PCR and cloning are shown in Supplementary information, Table S1.
Seeds were sterilized in 1% sodium hypochlorite and then washed five times using sterilized water. Vernalization was carried out in a 4 °C refrigerator for 2 days. Seedlings were grown on 1/2 MS medium plates containing 0.8% agar in a 16 h-light/8 h-dark and 22 °C growth chamber. For peptide treatment, synthetic sulfated RGF1 or synthetic sulfated FLAG-RGF1 was added to 1/2 MS medium. For liquid culture, 150-ml flasks containing 35 ml 1/2 MS medium and 35S::RGI1-FLAG transgenic seedlings were shaked at 120 rpm under the same growing condition.
RNA extraction, RT-PCR, and expression assay
Total RNA was extracted from Col-0, rgi1-1 rgi2-1 rgi3-1 rgi4-1 rgi5-1, rgi1-2 rgi2-2 rgi3-2 rgi4-2 rgi5-2, and pRGI2::gRGI2 (in rgi1-1 rgi2-1 rgi3-1 rgi4-1 rgi5-1) seedlings at 10 days after germination (DAG) using an RNA extraction kit (Tiangen, DP437). Four μg total RNA of each sample was used for synthesizing cDNA using an M-MLV kit (Invitrogen, C28025). Semi-quantitative RT-PCR was carried out to test the expression level of each allelic gene. The primers used for RT-PCR are shown in Supplementary information, Table S1.
For quantitative RT-PCR, the whole seedlings of Col-0, rgi1-1 rgi2-1 rgi3-1 rgi4-1 rgi5-1, and tpst-1 at 5 DAG were treated with 20 μM synthetic sulfated RGF1 for 3 h. RNA extraction and reverse transcription were carried out according to the aforementioned method. The primers used for quantitative RT-PCR are shown in Supplementary information, Table S1.
Staining and microscopy
Five-day-old seedlings of transgenic plants harboring pRGI1::GUS, pRGI2::GUS, pRGI3::GUS, pRGI4::GUS, and pRGI5::GUS were used for GUS staining for 30 min, as described previously32. Five-day-old seedlings of Col-0 and rgi1-1 rgi2-1 rgi3-1 rgi4-1 rgi5-1 were used for transverse root section assay after staining with toluidine blue. Photographs were taken with a microscopy (Leica, DM600B). For confocal imaging, seedlings were stained in 0.02 mg/ml PI for 12 min. CLSM was used for photographing (Leica, TCS SP8).
Protein purification, dot blotting, pull-down, and autophosphorylation assays
The extracellular domain of RGI1, not including the predicted signal peptide, was PCR amplified and cloned into a pMAL-cRI-GWR vector, and the resulting construct was transformed into E. coli Rosetta for protein expression. IPTG was added in a 50 ml liquid culture when OD researched 0.5, to make a final concentration of 0.2 mM, followed by a further incubation at 16 °C for 20 h. E. coli was collected through centrifugation at 5 000 rpm. for 10 min. The pellets were washed one time with amylase column buffer (20 mM Tris, 200 mM NaCl, 1 mM EDTA, pH7.5), suspended in 8 ml amylase column buffer with 1mM PMSF, and ultrasonically disrupted. The solution was centrifuged with 18 000× g at 4 °C for 30 min. The supernatant was incubated with prewashed amylase resin (NEB, E8021S) overnight. The resin was then washed five times by amylase column buffer and eluted using 10 mM maltose monohydrate in 10 mM Tris-HCl buffer, pH7.5. 100 ng MBP and MBP-RGI1ED was blotted on a nitrocellulose membrane, and blocked with 5% non-fat milk for 1 h at 4 °C. 20 μM FLAG-RGF1 in amylase column buffer was incubated with the membrane, and an anti-FLAG antibody was used as a primary antibody (AbMART, M20008) and anti-mouse IgG (H+L) (KPL) as a secondary antibody. Western Lightning Plus-ECL (Perkin Elmer) was used for chemiluminescence detection.
For pull-down analysis, resins associated MBP and MBP-RGI1ED were incubated with amylase column buffer containing 100 μM FLAG-RGF1 peptide at 4 °C for 1 h, and then washed four times with amylase column buffer. 2× SDS loading buffer was added and the mix was boiled for 5 min. The supernatant was used for Tris-Tricine-SDS-PAGE60 and analyzed with an anti-FLAG antibody.
The cytoplasmic domain (CD) of RGI1 was fused with MBP, and the fusion protein was expressed in E. coli as described above. After purification, 15 mM maltose was used to elute. The purified protein was analyzed by Coomassie Brilliant Blue staining and immunoblotting using an anti-pThr antibody (Cell Signaling, 9381S). The mutations in ATP-binding site of RGI1CD, K814E and K815E, were also analyzed.
Membrane protein isolation, immunoprecipitation, CIP treatment and western blotting
Ten-day-old transgenic seedlings harboring 35S::RGI1-FLAG cultured in 1/2 MS liquid medium were treated with mock or 20 μM RGF1 for 30 min. Ground to fine power in liquid N2 and membrane protein was extracted as described previously35. Anti-FLAG M2 Affinity Gel (Sigma, A2220) was used to purify RGI1-FLAG from membrane protein through immunoprecipitation. The immunopreciptated protein was analyzed by immunoblotting using an anti-FLAG antibody (AbMART, M20008) and an anti-pThr antibody after separation in SDS-PAGE. The immunopreciptated protein was also used for CIP (NEB, M0290V) treatment at 37 °C for 3 h, and then analyzed by an anti-FLAG antibody. An anti-ubiquitin (P4D1) antibody (Cell signaling, 3936S) was used to detect ubiquitination of RGI1.
Author Contributions
JL and YO designed the experiments. YO performed most of the experiments with the assistance from XL, QZ, QX, JZ, YW, HS, and ZW. JL and YO analyzed the data. BZ helped with the figures. CL and XZ provided materials. KH and XG participated in discussions. JL and YO wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
Matsubayashi Y . Posttranslationally modified small-peptide signals in plants. Annu Rev Plant Biol 2014; 65:385–413.
Grienenberger E, Fletcher JC . Polypeptide signaling molecules in plant development. Curr Opin Plant Biol 2015; 23:8–14.
Rehemtulla A, Kaufman RJ . Protein processing within the secretory pathway. Curr Opin Biotechnol 1992; 3:560–565.
Tabata R, Sumida K, Yoshii T, Ohyama K, Shinohara H, Matsubayashi Y . Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 2014; 346:343–346.
Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR . A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 2014; 343:408–411.
Wang J, Li H, Han Z, et al. Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature 2015; 525:265–268.
Betsuyaku S, Sawa S, Yamada M . The function of the CLE peptides in plant development and plant-microbe interactions. Arabidopsis Book 2011; 9:e0149.
Jun J, Fiume E, Roeder AH, et al. Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol 2010; 154:1721–1736.
Fiers M, Golemiec E, Xu J, et al. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 2005; 17:2542–2553.
Fiers M, Golemiec E, van der Schors R, et al. The CLAVATA3/ESR motif of CLAVATA3 is functionally independent from the nonconserved flanking sequences. Plant Physiol 2006; 141:1284–1292.
Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM . Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 1999; 283:1911–1914.
Kondo T, Sawa S, Kinoshita A, et al. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 2006; 313:845–848.
Clark SE, Williams RW, Meyerowitz EM . The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 1997; 89:575–585.
Kinoshita A, Betsuyaku S, Osakabe Y, et al. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 2010; 137:3911–3920.
Muller R, Bleckmann A, Simon R . The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 2008; 20:934–946.
Kayes JM, Clark SE . CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 1998; 125:3843–3851.
Zhu Y, Wang Y, Li R, et al. Analysis of interactions among the CLAVATA3 receptors reveals a direct interaction between CLAVATA2 and CORYNE in Arabidopsis. Plant J 2010; 61:223–233.
Bleckmann A, Weidtkamp-Peters S, Seidel CA, Simon R . Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol 2010; 152:166–176.
Stahl Y, Wink RH, Ingram GC, Simon R . A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol 2009; 19:909–914.
Stahl Y, Grabowski S, Bleckmann A, et al. Moderation of Arabidopsis root stemness by CLAVATA1 and Arabidopsis CRINKLY4 receptor kinase complexes. Curr Biol 2013; 23:362–371.
Fisher K, Turner S . PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr Biol 2007; 17:1061–1066.
Hirakawa Y, Shinohara H, Kondo Y, et al. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc Natl Acad Sci USA 2008; 105:15208–15213.
Whitford R, Fernandez A, De Groodt R, Ortega E, Hilson P . Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc Natl Acad Sci USA 2008; 105:18625–18630.
Etchells JP, Turner SR . The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 2010; 137:767–774.
Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y . Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 2010; 329:1065–1067.
Meng L, Buchanan BB, Feldman LJ, Luan S . CLE-like (CLEL) peptides control the pattern of root growth and lateral root development in Arabidopsis. Proc Natl Acad Sci USA 2012; 109:1760–1765.
Whitford R, Fernandez A, Tejos R, et al. GOLVEN secretory peptides regulate auxin carrier turnover during plant gravitropic responses. Dev Cell 2012; 22:678–685.
Fernandez A, Drozdzecki A, Hoogewijs K, et al. Transcriptional and functional classification of the GOLVEN/ROOT GROWTH FACTOR/CLE-like signaling peptides reveals their role in lateral root and hair formation. Plant Physiol 2013; 161:954–970.
Komori R, Amano Y, Ogawa-Ohnishi M, Matsubayashi Y . Identification of tyrosylprotein sulfotransferase in Arabidopsis. Proc Natl Acad Sci USA 2009; 106:15067–15072.
Zhou W, Wei L, Xu J, et al. Arabidopsis Tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. Plant Cell 2010; 22:3692–3709.
Shiu SH, Bleecker AB . Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 2001; 98:10763–10768.
Wu Y, Xun Q, Guo Y, et al. Genome-wide expression pattern analyses of the Arabidopsis leucine-rich repeat receptor-like kinases. Mol Plant 2016; 9:289–300.
Gou X, Yin H, He K, et al. Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet 2012; 8:e1002452.
Du J, Yin H, Zhang S, et al. Somatic embryogenesis receptor kinases control root development mainly via brassinosteroid-independent actions in Arabidopsis thaliana. J Integr Plant Biol 2012; 54:388–399.
Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC . BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 2002; 110:213–222.
Nam KH, Li J . BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 2002; 110:203–212.
Wang X, Kota U, He K, et al. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell 2008; 15:220–235.
Li J . Multi-tasking of somatic embryogenesis receptor-like protein kinases. Curr Opin Plant Biol 2010; 13:509–514.
Santiago J, Henzler C, Hothorn M . Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 2013; 341:889–892.
Sun Y, Han Z, Tang J, et al. Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res 2013; 23:1326–1329.
Chinchilla D, Zipfel C, Robatzek S, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 2007; 448:497–500.
Heese A, Hann DR, Gimenez-Ibanez S, et al. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 2007; 104:12217–12222.
Sun Y, Li L, Macho AP, et al. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 2013; 342:624–628.
Gou X, He K, Yang H, et al. Genome-wide cloning and sequence analysis of leucine-rich repeat receptor-like protein kinase genes in Arabidopsis thaliana. BMC Genomics 2010; 11:19.
Obrdlik P, El-Bakkoury M, Hamacher T, et al. K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc Natl Acad Sci USA 2004; 101:12242–12247.
Di Laurenzio L, Wysocka-Diller J, Malamy JE, et al. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 1996; 86:423–433.
Helariutta Y, Fukaki H, Wysocka-Diller J, et al. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 2000; 101:555–567.
Nakajima K, Sena G, Nawy T, Benfey PN . Intercellular movement of the putative transcription factor SHR in root patterning. Nature 2001; 413:307–311.
Sabatini S, Heidstra R, Wildwater M, Scheres B . SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 2003; 17:354–358.
Wildwater M, Campilho A, Perez-Perez JM, et al. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 2005; 123:1337–1349.
Aida M, Beis D, Heidstra R, et al. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 2004; 119:109–120.
Galinha C, Hofhuis H, Luijten M, et al. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 2007; 449:1053–1057.
Fernandez A, Drozdzecki A, Hoogewijs K, et al. The GLV6/RGF8/CLEL2 peptide regulates early pericycle divisions during lateral root initiation. J Exp Bot 2015; 66:5245–5256.
Horn MA, Walker JC . Biochemical properties of the autophosphorylation of RLK5, a receptor-like protein kinase from Arabidopsis thaliana. Biochim Biophys Acta 1994; 1208:65–74.
Shinohara H, Mori A, Yasue N, Sumida K, Matsubayashi Y . Identification of three LRR-RKs involved in perception of root meristem growth factor in Arabidopsis. Proc Natl Acad Sci USA 2016; 113:3897–3902.
Song W, Liu L, Wang J, et al. Signature motif-guided identification of peptide hormone receptors regulating root meristem growth. Cell Res 2016 May 27. doi: 10.1038/cr.2016.62
Li J, Chory J . A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 1997; 90:929–938.
Wang X, Goshe MB, Soderblom EJ, et al. Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 2005; 17:1685–1703.
Clough SJ, Bent AF . Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 1998; 16:735–743.
Schagger H . Tricine-SDS-PAGE. Nat Protoc 2006; 1:16–22.
Acknowledgements
We are grateful to the Arabidopsis Biological Resource Center for the T-DNA insertion lines used in these studies. We also thank Wendy Chen for reading and commenting on the manuscript and Zeping Cai for providing transgenic plant seeds carrying pSHR::NLS-YFP, pSCR::NLS-YFP, or pWOX5::NLS-YFP. This study was supported by the National Natural Science Foundation of China (31470380, 31530005, and 91317311 to JL).
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
Expression patterns of five RGIs, confirmation of their T-DNA insertions, and the phenotypes of rgi1-1 rgi2-1 rgi3-1 rgi4-1 rgi5-1 shown in root cross section and aerial parts. (PDF 611 kb)
Supplementary information, Figure S2
Confirmation of two independent lines of transgenic plants, harboring pRGI2::gRGI2 in rig1,2,3,4,5 by RT-PCR analysis. (PDF 356 kb)
Supplementary information, Figure S3
Expression patterns of several marker genes critical for root development and phenotypes of several mutants. (PDF 533 kb)
Supplementary information, Figure S4
The rig1,2,3,4,5 quintuple mutant showed a phenotype similar to the null alleles of the components in RGF1-PLT pathway. (PDF 599 kb)
Supplementary information, Figure S5
The expression levels of PLT1 and PLT2 cannot be induced in rig1,2,3,4,5 but can be induced in Col-0 and tpst-1 by RGF1 treatment. (PDF 369 kb)
Supplementary information, Figure S6
Subcellular localization of RGI1, biological activity assay of synthetic sulfated FLAG-RGF1, and pull-down analysis of FLAG-RGF1 and RGI1. (PDF 515 kb)
Supplementary information, Table S1
Primers used in this work (PDF 434 kb)
Rights and permissions
About this article
Cite this article
Ou, Y., Lu, X., Zi, Q. et al. RGF1 INSENSITIVE 1 to 5, a group of LRR receptor-like kinases, are essential for the perception of root meristem growth factor 1 in Arabidopsis thaliana. Cell Res 26, 686–698 (2016). https://doi.org/10.1038/cr.2016.63
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2016.63
Keywords
This article is cited by
-
The LRR receptor-like kinase ALR1 is a plant aluminum ion sensor
Cell Research (2024)
-
Mobile Signaling Peptides: Secret Molecular Messengers with a Mighty Role in Plant Life
Journal of Plant Growth Regulation (2023)
-
CLE peptides: critical regulators for stem cell maintenance in plants
Planta (2022)
-
Transcriptome analysis revealed the regulation of gibberellin and the establishment of photosynthetic system promote rapid seed germination and early growth of seedling in pearl millet
Biotechnology for Biofuels (2021)
-
Identification of tomato root growth regulatory genes and transcription factors through comparative transcriptomic profiling of different tissues
Physiology and Molecular Biology of Plants (2021)