Abstract
The eukaryotic genome is organized into chromatins, the physiological template for DNA-dependent processes including replication, recombination, repair, and transcription. Chromatin-mediated transcription regulation involves DNA methylation, chromatin remodeling, and histone modifications. However, chromatin also contains non-histone chromatin-associated proteins, of which the high-mobility group (HMG) proteins are the most abundant. Although it is known that HMG proteins induce structural changes of chromatin, the processes underlying transcription regulation by HMG proteins are poorly understood. Here we decipher the molecular mechanism of transcription regulation mediated by the HMG AT-hook 2 protein (HMGA2). We combined proteomic, ChIP-seq, and transcriptome data to show that HMGA2-induced transcription requires phosphorylation of the histone variant H2AX at S139 (H2AXS139ph; γ-H2AX) mediated by the protein kinase ataxia telangiectasia mutated (ATM). Furthermore, we demonstrate the biological relevance of this mechanism within the context of TGFβ1 signaling. The interplay between HMGA2, ATM, and H2AX is a novel mechanism of transcription initiation. Our results link H2AXS139ph to transcription, assigning a new function for this DNA damage marker. Controlled chromatin opening during transcription may involve intermediates with DNA breaks that may require mechanisms that ensure the integrity of the genome.
Similar content being viewed by others
Introduction
The nucleosome is the structural and functional unit of chromatin and consists of a nucleosome core particle, a linker histone H1, and linker DNA1. The nucleosome core particle is built up 147 bp of DNA wrapped around a histone octamer, which is composed of two copies of each of the four core histones (H2A, H2B, H3, and H4). In addition to nucleosomes, chromatin consists of non-histone chromatin-associated proteins, of which the high-mobility group (HMG) proteins are the most abundant. Based on their DNA-binding domains, the HMG proteins are subdivided into three families: HMGA (containing AT-hooks), HMGB (containing HMG-boxes), and HMGN (containing nucleosomal-binding domains)2,3. HMG proteins bind to specific structures in DNA or chromatin in a sequence-independent manner via their respective functional motifs4. Interestingly, although they do not possess intrinsic transcriptional activity, HMG proteins are considered as architectural transcription factors because they modulate transcription of their target genes by altering the chromatin structure at the promoter and/or enhancers4. Here we deciphered the mechanism of transcriptional regulation mediated by HMGA2, an HMGA family member that mediates transforming growth factor β 1 (TGFβ1) signaling5.
The serine/threonine protein kinase ATM belongs to the superfamily of phosphatidylinositol 3-kinase-related kinases. Mutations in the ATM gene cause a rare autosomal recessive neurodegenerative disorder named ataxia telangiectasia, which involves a marked defect in DNA double-strand break (DSB) repair6. A well-known substrate of ATM is the histone variant H2AX, which represents about 2%-25% of the cellular H2A pool in mammals7. ATM phosphorylates H2AX on serine 139. Phosphorylated H2AX at S139 (H2AXS139ph, γ-H2AX) is often used as a marker for DNA-DSB8. However, accumulating evidences suggest additional functions of this histone modification9,10,11. In addition, McManus et al.12 analyzed the dynamics of H2AXS139ph in normal growing mammalian cells and found ATM-dependent H2AX phosphorylation in all phases of cell cycle with a maximum during M phase, suggesting that H2AXS139ph may contribute to the fidelity of the mitotic process, even in the absence of DNA damage, thereby ensuring the correct transmission of genetic information from one generation to the next. In this study, we report for the first time that H2AXS139ph plays a role in HMGA2-induced transcriptional activation.
Results
HMGA2, ATM, and H2AXS139ph transcriptional network
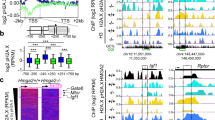
A high-resolution mass spectrometry-based proteomic approach to identify HMGA2-interacting proteins (Figure 1A) and subsequent confirmation by co-immunoprecipitation (Co-IP) of endogenous proteins (Figure 1B) revealed that HMGA2 interacts with the protein kinase ATM, the histone variant H2AX and its phosphorylated form H2AXS139ph. Significant interaction was not detected between HMGA2 and other members of the superfamily of phosphatidylinositol 3-kinase-related kinases, ATR or PRKDC. Thus, we decided to focus our attention on a potential role of ATM-mediated protein phosphorylation during transcription regulated by HMGA2. We performed a chromatin immunoprecipitation sequencing (ChIP-seq) experiment (Figure 1C-1F and Supplementary information, Figure S1 and S2) using HMGA2-, ATM-, and H2AXS139ph-specific antibodies (Supplementary information, Figure S1A) and chromatin isolated from mouse embryonic fibroblasts (MEF) from wild-type (WT) and Hmga2-deficient (Hmga2−/−)13 embryos. We analyzed the peaks obtained from WT MEF (Figure 1C, left) and observed 63.1% (P < 0.001) common target genes among HMGA2, ATM, and H2AX139ph. KEGG pathway-enrichment analysis (right) showed that these common target genes are related to cell adhesion, TGFβ, and WNT signaling. Further analysis showed significant co-occupancy of ATM and H2AXS139ph at HMGA2-binding sites (Figure 1D), correlating with our HMGA2 interactome data. We decided to investigate whether Hmga2 was required for the genome-wide-binding profile of H2AXS139ph (Figure 1E). Since we hypothesized a mechanism of transcriptional activation, we focused our analysis on promoters. When compared with WT MEF, Hmga2-knockout (KO) significantly reduced the number of genes with H2AXS139ph peaks at the promoters from 7.5% to 4.7% (P < 0.0001). Accordingly, ATM and H2AXS139ph occupancy at the promoters was dramatically reduced after Hmga2-KO (Figure 1F), demonstrating the requirement of HMGA2 for the genome-wide promoter-binding profile of these two proteins and suggesting the possibility of an HMGA2-mediated ATM recruitment to the promoters and subsequent phosphorylation of H2AX. Visualization of different gene loci using UCSC genome browser supported these observations (Figure S2A). Strikingly, ChIP assay in WT MEF showed enrichment of HMGA2, ATM, and H2AXS139ph at the promoters of Gata6, Snai1, and Snai2 (Figure 1G), three of the genes identified in our ChIP-seq experiment. Moreover, Hmga2-KO significantly reduced the binding of these three proteins at the analyzed promoters, confirming our ChIP-seq results. Affymetrix microarray-based transcriptome analysis (Supplementary information, Figure S2B) showed that Hmga2-KO reduced the expression of genes with co-occupancy of HMGA2, ATM, and H2AXS139ph in WT MEF, supporting a mechanism of transcription that requires HMGA2 and involves ATM-mediated H2AX phosphorylation.
HMGA2, ATM, and H2AXS139ph transcriptional network. (A) HMGA2-interacting proteins identified by mass spectrometry analysis. SILAC ratios were derived from label free quantitation between HMGA2 and control co-affinity precipitations. Scatter plot between SILAC ratios and peak intensities (left); selected proteins with corresponding log2 SILAC ratios (right). (B) Confirmation of proteomic data. Endogenous HMGA2 was immunoprecipitated (IP) from nuclear protein extracts of MLE-12 cells using either immunoglobulin G (IgG, as control) or HMGA2-specific antibody. Co-IP proteins were analyzed by WB using the indicated antibodies. Input, 5% of material used for the IP; H2AXS139ph, H2AX phosphorylated at serine 139. (C) Venn diagram displaying % overlap of common target genes among HMGA2 (blue), H2AXS139ph (green), and ATM (deeppink) after ChIP-seq in wild-type (WT) MEF (left; P < 0.001). KEGG pathway-enrichment analysis (right) using DAVID bioinformatics tool (P < 0.01 and FDR < 4). j, junction; ad, adhesion. (D) Aggregate plot for ATM and H2AXS139ph peaks in a ± 2 kb region respective to HMGA2 peaks. Norm tags, tag density normalized to 3 × 107 using annotatePeaks.pl from HOMER. (E) Genome-wide distribution of H2AXS139ph peaks relative to gene structure in WT (Hmga2+/+) and Hmga2−/− MEF (P < 0.0001). Square on right, enlargement of the graph. Prom, promoter; TTS, transcription termination site. (F) Aggregate plot for ATM and H2AXS139ph peaks relative to promoter (prom) region in Hmga2+/+ (black) and Hmga2−/− (gray) MEF. Data were processed with ngs.plot using bed files of each. Values are shown as log2 and normalized to input. (G) HMGA2 is required for H2AXS139ph and ATM association on Gata6, Snai1, and Snai2 promoters. ChIP of Gata6, Snai1, and Snai2 promoters after HMGA2-, H2AXS139ph-, and ATM-IP in Hmga2+/+ and Hmga2−/− MEF. In all figures, error bars, sem (n = 3); asterisks, P values after one-way ANOVA, ***P < 0.001; **P < 0.01; *P < 0.05.
TGFβ1-induced transcription requires HMGA2, ATM, and H2AXS139ph
HMGA2 mediates TGFβ1 signaling5. Thus, we analyzed whether the mechanism of transcriptional activation proposed here might play a role within the biological context of TGFβ1 signaling. Western blot (WB) analysis of protein extracts from MEF (Figure 2A) and normal murine mammary gland (NMuMG) epithelial cells (Figure 2B) showed increased levels of H2AXS139ph after TGFβ1 treatment without affecting the total levels of H2AX, indicating that the observed effects were indeed related to the post-translational modification. Induction of TGFβ1 signaling in both cell types was monitored by immunodetection of phosphorylated SMAD3 (pSMAD3). Interestingly, Hmga2-KO not only reduced the basal H2AXS139ph levels in MEF (Figure 2A, lane 3) but also blocked the effects of TGFβ1 treatment on this histone modification (lane 4). Moreover, Atm-specific loss of function (LOF) using a small interfering RNA (siRNA; siAtm) in NMuMG cells (Figure 2B, lane 2) abrogated the inductive effect of TGFβ1 on H2AXS139ph levels (lane 4).
TGFβ1-induced transcription requires HMGA2, ATM, and H2AXS139ph. (A) TGFβ1 induces H2AXS139ph in an HMGA2-dependent manner. Protein extracts from Hmga2+/+ and Hmga2−/− MEF that were either untreated (−) or treated (+) with TGFβ1 were analyzed by WB using the indicated antibodies. pSMAD3, phosphorylated SMAD3, a marker for TGFβ1 signaling; ACTB, β actin, as loading control. (B) TGFβ1 induces H2AXS139ph in an ATM-dependent manner. Protein extracts from NMuMG cells that were transiently transfected with control (siCtrl) or Atm-specific small interfering RNA (siAtm) and treated with TGFβ1 were analyzed as in A. (C) TGFβ1 induces H2AX phosphorylation and enrichment of active RNA polymerase II (Pol II) on Gata6, Snai1, and Snai2 promoters in an HMGA2- and ATM-dependent manner. ChIP on the Gata6, Snai1, and Snai2 promoters using the indicated antibodies was performed in MLE-12 cells that were transiently transfected with siCtrl, siHmga2, or siAtm, or treated with ATM inhibitor KU-55933 (ATMi) prior to TGFβ1 treatment as indicated. Pol II, carboxy-terminal domain phosphorylated Pol II. (D) HMGA2 is required for TGFβ1-induced expression of Gata6, Snai1, and Snai2. Quantitative RT-PCR-based gene expression analysis in MLE-12 cells that were stably transfected with Ctrl or Hmga2-specific shDNA and treated with TGFβ1 as indicated. Rel nor exp, relative expression normalized to Gapdh.
To explore the effect of TGFβ1 on H2AXS139ph levels at specific loci, we examined the promoters of Gata6, Snai1, and Snai2 in MLE-12 cells by ChIP (Figure 2C), and detected an increase of H2AXS139ph at these promoters following TGFβ1 treatment (Figure 2C, top). Interestingly, expression of siRNA-targeting Hmga2 (siHmga2), or Atm (siAtm), or treatment with ATM inhibitor KU-5593314 (ATMi; Supplementary information, Figure S3A) abrogated the effect of TGFβ1 treatment, confirming our previous WB results. In addition, TGFβ1 treatment led to accumulation of HMGA2, ATM, pSMAD3, and transcription initiating S5 phosphorylated RNA polymerase II (Pol II) on all three promoters in an Hmga2- and Atm-dependent manner (Figure 2C, middle and bottom). Moreover, TGFβ1-induced accumulation of HMGA2 was reduced when ATM was disrupted, suggesting that ATM also controls the recruitment of HMGA2 to TGF-responsive genes. Consistent with our ChIP analysis, we detected increased levels of Gata6, Snai1, and Snai2 transcripts after TGFβ1 treatment in an Hmga2-dependent manner (Figure 2D) as shown by quantitative RT-PCR (qRT-PCR)-based expression analysis in MLE-12 cells that were stably transfected either with a control (Ctrl) or an Hmga2-specific short hairpin DNA (shDNA) construct. Our results demonstrate that TGFβ1-induced transcriptional activation involves H2AXS139ph and requires HMGA2 and ATM.
TGFβ1 inhibits cell cycle progression15, while HMGA2 acts as either a cell cycle activator or an inhibitor depending on the cellular context5,16,17. In addition, several reports suggest an increase of H2AXS139ph during cell cycle progression due to DNA replication stress18,19. Thus, to discriminate between transcription-related H2AXS139ph and DNA replication stress-induced H2AXS139ph, we used L-mimosine, a cell cycle inhibitor that is known to increase DNA replication stress20,21. Protein extracts of MLE-12 cells that were either transfected with Hmga2 or treated with L-mimosine or TGFβ1 were analyzed by WB (Supplementary information, Figure S3C). Induction of TGFβ1 signaling was monitored by immunodetection of phosphorylated SMAD2 (pSMAD2). As expected, L-mimosine and TGFβ1 treatment decreased the levels of cyclin A2 (CCNA2) indicating cell cycle arrest15,20,21. Hmga2 transfection minimally reduced CCNA2 levels. In addition, all three treatments increased the levels of H2AXS139ph when compared with the control transfected cells. Interestingly, only Hmga2 transfection and TGFβ1 treatment increased the levels of GATA6, demonstrating a novel function of H2AXS139ph during transcriptional activation that is independent of DNA replication stress after cell cycle arrest.
HMGA2 activates transcription via H2AX phosphorylation
We have previously shown that HMGA2 directly regulates Gata616. Thus, we focused on the Gata6 promoter to further elucidate the mechanism of transcriptional activation. In addition to the Hmga2−/− mice13, we used a second mouse line expressing Hmga2 with a truncated 3′ UTR that stabilizes the transcript resulting in higher levels of HMGA222, thereby creating transgenic gain-of-function (GOF) conditions when compared to WT mice. WB of protein extracts from mouse embryos (Figure 3A) showed that H2AXS139ph levels decreased after Hmga2-KO (lanes 2 and 3), confirming our results in MEF (Figure 2A), whereas transgenic Hmga2-GOF elevated H2AXS139ph levels. (lanes 5 and 6). Interestingly, ChIP revealed similar results at the Gata6 promoter (Figure 3B); H2AXS139ph levels decreased after Hmga2-KO, whereas increased after Hmga2-GOF. These results were confirmed by sequential ChIP (ChIP-reChIP) using an antibody specific for phosphorylated ATM substrates (ATM subs) and subsequently H2AX antibody (Supplementary information, Figure S4). Consistent with our in vitro data (Figure 2C), transgenic Hmga2-GOF increased the association of HMGA2, ATM, and Pol II to the Gata6 promoter when compared with WT mice (Figure 3C), whereas Hmga2-KO induced the reverse effect. Moreover, expression analysis in mouse embryos (Figure 3D) showed 50% decrease of Gata6 expression after Hmga2-KO and more than fivefold increase after Hmga2-GOF when compared with WT mice, correlating with our ChIP results.
HMGA2 activates transcription via H2AX phosphorylation. (A) HMGA2 modulates H2AX phosphorylation at S139. Protein extracts from Hmga2+/+, Hmga2−/−, and transgenic Hmga2 gain-of-function (GOF) mouse embryos (E18.5) were analyzed by WB using the indicated antibodies. Two embryos per transgenic mouse line were analyzed. LMNB1, Lamin B1, as loading control. (B) Hmga2 is required for H2AXS139ph enrichment on Gata6 promoter. ChIP analysis of the Gata6 promoter in Hmga2+/+, Hmga2−/−, and Hmga2-GOF E18.5 embryonic lungs using the indicated antibodies. IgG, negative control. (C) Hmga2-knockout (KO) reduces, whereas Hmga2 GOF increases, the levels of ATM and Pol II on the Gata6 promoter. ChIP analysis as in B. (D) Hmga2-KO reduces, whereas Hmga2 GOF increases, Gata6 expression. qRT-PCR-based Gata6 expression analysis in Hmga2+/+, Hmga2−/−, and Hmga2 GOF E18.5 embryonic lungs. Rel nor exp, relative expression normalized to Tuba1a. (E) Hmga2 is essential for ATM-mediated H2AX phosphorylation. Nuclear protein extracts from Hmga2+/+, Hmga2−/−, and Hmga2 GOF mice lung were analyzed by IP using either an antibody specific for phosphorylated ATM substrate (ATM subs) or IgG as control. Immunoprecipitated proteins were analyzed by WB using the indicated antibodies. Input, 5% of material used for the IP. (F) HMGA2 is required for ATM-mediated H2AX phosphorylation. HEK293T cells stably expressing H2AX-flag were transiently transfected with an empty vector (−) or an Hmga2-myc expression plasmid and control (siCtrl) or HMGA2-specific siRNA (siHMGA2). Nuclear protein extracts of transfected cells were analyzed as in E. (G) HMGA2 is essential for the interaction of ATM with H2AX. HEK293T cells stably expressing H2AX-flag were transfected with either control (siCtrl) or HMGA2-specific siRNA (siHMGA2). Nuclear protein extracts of transfected cells were analyzed by IP using ATM-specific antibody or IgG as control. Immunoprecipitated proteins were analyzed by WB using the indicated antibodies. All graphs show error bars, sem (n = 4) and asterisks as in Figure 1G.
Co-IP using the ATM subs antibody (Figure 3E) showed that Hmga2-KO reduced H2AX-phosphorylation (lane 2), whereas transgenic Hmga2-GOF induced the opposite effect (lane 3). These observations were in line with the results that we obtained from HEK293T cells (Figure 3F) where siRNA-mediated HMGA2-LOF inhibited H2AX-phosphorylation (lane 3). The specificity of this finding was confirmed by a partial rescue of H2AX-phosphorylation in HMGA2-depleted HEK293T cells after expression of the mouse Hmga2 ortholog (lane 6). In addition, HMGA2-LOF blocked the interactions between ATM and the histones H2AX (Figure 3G; lane 2), suggesting that HMGA2 mediates the molecular interaction between ATM and H2AX.
ATM is required for HMGA2-mediated transcriptional activation
To show the requirement of ATM for the mechanism of transcription proposed here, HEK293T cells were transiently transfected with Hmga2 and a plasmid containing the luciferase (Luc) reporter gene under the control of the Gata6 promoter (Figure 4A, top). ATM-LOF was induced either by siRNA transfection (siATM) or ATMi treatment (Supplementary information, Figure S3A and S3B). Forced expression of Hmga2 increased the basal transcription of the Gata6-Luc reporter by more than fourfold. In addition, expression of endogenous GATA6 increased more than threefold after Hmga2 transfection (bottom). Interestingly, both ATM-LOF approaches counteracted the enhanced expression of the Gata6-Luc reporter as well as endogenous GATA6 after transfection of Hmga2. Since the Gata6-Luc reporter used contains only the Gata6 promoter, our results support a mechanism of transcription initiation at the promoter. WB of protein extracts from HEK293T cells (Figure 4B) showed increased levels of GATA6 after Hmga2-GOF (lane 2) that were reduced after ATMi treatment (lane 3) supporting our expression analysis. Similarly, transfection of Hmga2-myc increased the levels of H2AXS139ph (lane 2) in an ATM-dependent manner (lane 3). Neither Hmga2 transfection nor ATMi altered the total level of H2AX, showing again that the observed effect is related to post-translational modification of H2AX. Furthermore, we analyzed the GATA6 promoter in HEK293T cells by ChIP after transfection of empty vector (Ctrl) or Hmga2 (Figure 4C). Hmga2-GOF led to an enrichment of endogenous ATM, H2AXS139ph, and Pol II at the GATA6 promoter. Interestingly, ATM-LOF antagonized the effects caused by Hmga2 transfection, demonstrating the requirement of ATM for these effects. The increase of H2AXS139ph after Hmga2-GOF in ATM-dependent manner was confirmed by immunostaining in HEK293T cells (Supplementary information, Figure S3D).
HMGA2-induced transcription requires ATM. (A) ATM-LOF counteracted Hmga2-induced transcriptional activation of GATA6. Top: luciferase reporter assays of HEK293T cells that were transiently transfected with a Gata6-luciferase (Gata6-Luc) reporter plasmid and either an empty vector (Ctrl) or an Hmga2 expression plasmid. These cells were further transfected with either and siCtrl or siATM, or treated with ATM inhibitor KU-55933 (ATMi). RLU, relative light units. Bottom: endogenous GATA6 expression monitored by qRT-PCR in HEK293T cells. Rel nor exp, relative expression normalized to Tuba1a. (B) Inhibition of ATM kinase abrogates the effect of Hmga2 on GATA6 levels. HEK293T cells were transiently transfected with either an empty vector (−) or an Hmga2-myc expression plasmid. Transfected cells were either untreated (−) or treated with ATMi. Nuclear protein extracts were analyzed by WB using the indicated antibodies. (C) ATM-LOF antagonized Hmga2-induced enrichment of ATM, H2AXS139ph, and Pol II on GATA6 promoter. ChIP analysis was performed using the indicated antibodies in HEK293T cells treated as in A. (D) Hmga2-induced GATA6 increment involves DNA damage-independent H2AXS139ph. HEK293T cells were transiently transfected with Ctrl or an Hmga2 expression plasmid or treated with UV-C or H2O2. Nuclear protein extracts were analyzed by WB using the indicated antibodies. TRP53BP1 (transformation-related protein 53-binding protein 1) and RAD51 (RAD51 recombinase homolog) serve as DNA damage markers. All graphs show error bars, sem (n = 3) and asterisks as in Figure 1G.
H2AXS139ph is a marker for DNA-DSB8. To discriminate between the different roles of H2AXS139ph, nuclear protein extracts of HEK293T cells that were either transfected with Hmga2 or treated with UV or H2O2 were analyzed by WB (Figure 4D). All three types of treatments increased the levels of H2AXS139ph when compared with the control transfected cells. As expected, UV and H2O2 treatment increased the levels of the DNA repair proteins TRP53BP123,24 and RAD5125,26,27, indicating the presence of DNA damage and supporting the use of H2AXS139ph as marker for DNA-DSB under these conditions. Interestingly, while Hmga2 transfection did not change the levels of DNA repair proteins, it increased the levels of GATA6, demonstrating a novel function of H2AXS139ph during transcriptional activation.
H2AX phosphorylation is required for HMGA2-induced transcription
We created HEK293T cells that stably expressed a flag-tagged WT H2AX (H2AXWT) or a mutant H2AX (H2AXS-A), in which serine 139 was mutated to alanine (Figure 5A). Both H2AXwt and H2AXS-A immunoprecipitated along with ATM in Co-IP analysis of protein extracts using HMGA2-specific antibody (lanes 2-3), showing that the interaction between HMGA2, H2AX, and ATM was apparently not affected by the mutation. These results were confirmed by Co-IP using ATM-specific antibody (Supplementary information, Figure S5A). However, Co-IP using the ATM subs antibody (Figure 5B) showed that the S139A mutation abrogated the phosphorylation of H2AX (lane 3), demonstrating the requirement of this amino-acid residue for this specific post-translational modification. These results were confirmed by WB of protein extracts from both cell lines using H2AXS139ph-specific antibody (Supplementary information, Figure S5B). ChIP of chromatin from both cell lines (Figure 5C) showed accumulation of HMGA2, ATM, and Pol II on the endogenous GATA6 promoter after Hmga2 transfection that was more prominent in the cells expressing H2AXwt. The presence of H2AXS-A reduced the effects of Hmga2-GOF, suggesting that ATM-mediated H2AXS139ph is required for the HMGA2-induced changes in the chromatin of the GATA6 promoter. Moreover, expression analysis of endogenous GATA6 (Figure 5D) and a Gata6-Luc reporter (Supplementary information, Figure S5C) demonstrated enhanced GATA6 expression after Hmga2 transfection only in cells expressing H2AXwt, correlating with our ChIP results. WB of protein extracts from both HEK293T cell lines (Figure 5E) showed increased levels of GATA6 after Hmga2-GOF only in an H2AXwt background (lane 2), supporting our expression analysis. To further confirm the causal involvement of H2AXS139ph in the HMGA2-mediated transcription, we created a phosphomimetic H2AX mutant by substituting S139 with aspartic acid (H2AXS-D). Transfection of HEK293T with H2AXS-D (Figure 5F) increased GATA6 expression modestly. The increase of GATA6 expression by this phosphomimetic mutant became more significant when Hmga2 was co-transfected. Interestingly, the inhibiting effect of ATMi on Hmga2-enhanced GATA6 expression was partially overcome by H2AXS-D transfection, supporting that ATM-catalyzed H2AXS139ph mediates the enhancing effect of Hmga2 on GATA6 expression.
HMGA2-induced transcription requires H2AXS139ph. (A) Interaction of HMGA2 with ATM and H2AX is independent of H2AX phosphorylation site at serine 139. Nuclear protein extracts from HEK293T cells stably expressing flag-tagged wild-type (WT) or mutant (S-A, serine 139 to alanine) H2AX were analyzed by IP using an HMGA2-specific antibody or IgG as control. Immunoprecipitated proteins were analyzed by WB using the indicated antibodies. (B) S139A mutation (S-A) in H2AX abolishes ATM-mediated H2AX phosphorylation. Nuclear protein extracts from HEK293T cells were analyzed as in A using for IP an antibody specific for phosphorylated ATM substrates (ATM subs) or IgG as control. (C) H2AX S-A mutant blocks Hmga2-induced Pol II enrichment on GATA6 promoter. HEK293T cells stably expressing either WT or S-A mutant H2AX were transiently transfected with either an empty vector (Ctrl) or Hmga2. Chromatin was isolated 48 h after transfection and analyzed by ChIP using the indicated antibodies. (D) H2AX S-A mutant abolished Hmga2-induced transcriptional activation of GATA6. GATA6 expression was monitored by qRT-PCR in HEK293T cells that were transfected as in C. Rel nor exp, relative expression normalized to Tuba1a. (E) H2AX S-A mutant blocks the enhancing effect of Hmga2 on GATA6 levels. Nuclear protein extracts from HEK293T cells that were transfected as in D were analyzed by WB using the indicated antibodies. (F) The phosphomimetic mutant H2AX S139D (H2AXS-D) overcomes the effect of ATMi and partially rescues the Hmga2-induced transcriptional activation of GATA6. HEK293T cells that were treated with DMSO (−) or ATMi were transiently transfected with either an empty vector (Ctrl) or Hmga2 and/or H2AXS-D as indicated. GATA6 expression was monitored as in D. All graphs show error bars, sem (n = 3) and asterisks as in Figure 1G.
To reposition our mechanism of transcriptional activation into a biological context, we monitored epithelial-mesenchymal transition (EMT) after TGFβ1 treatment in NMuMG cells (Figure 6A). TGFβ1 potently induced EMT as shown by the loss of CDH1 (cadherin 1), an epithelial cell marker, and the gain of ACTA2 (alpha 2 smooth muscle actin), a mesenchymal cell marker. Interestingly, ATMi antagonized the effect caused by TGFβ1, showing the requirement of the kinase for TGFβ1-induced EMT. Transfection of the phosphomimetic mutant H2AXS-D alone was not sufficient to induce EMT, since it required TGFβ1 treatment. However, transfection of H2AXS-D reverted partially the repressive effect of ATMi, showing the causal involvement of ATM-mediated H2AXS139ph and demonstrating the biological relevance of the mechanism of transcriptional activation proposed here during TGFβ1-induced EMT. These results were confirmed by qRT-PCR-based expression analysis (Figure 6B) of the epithelial marker Cdh1, the mesenchymal marker Acta2, and the TGFβ1 target genes and EMT mediators Snai1 and Snai2.
(A) H2AXS-D overcomes the inhibiting effect of ATMi and partially rescues the TGFβ1-induced EMT. NMuMG cells were transiently transfected with either an empty vector (Ctrl) or H2AXS-D or/and treated with ATMi or TGFβ1 as indicated. Fluorescence microscopy of transfected/treated cells after double immunostaining using CDH1- and ACTA2-specific antibodies. Cell were also stained with DAPI for nucleus (blue). Scale bar, 40 μm. (B) quantitative RT-PCR-based expression analysis of the indicated genes in NMuMG cells transfected/treated as in A. Error bars, sem (n = 3) and asterisks as in Figure 1G. (C) Model: TGFβ1-induced transcription requires HMGA2 (blue hexagon) and ATM (deppink oval)-mediated phosphorylation (green circle) of H2AX.
Discussion
We combined proteomic-based interactome, ChIP-seq, and microarray-based transcriptome data to identify a novel transcriptional activation mechanism, which plays an important role in TGFβ1-signaling pathway. This mechanism of transcriptional activation requires HMGA2 and ATM-mediated H2AXS139ph (Figure 6C). Our results link H2AXS139ph, a marker for DNA damage, to transcriptional activation, which is an entirely new function for this histone modification. Furthermore, we demonstrate the relevance of this mechanism within the context of EMT, a process that is crucial during normal biological conditions as well as in certain diseases3.
H2AXS139ph is perhaps one of the most widely used biomarkers for DNA damage8. However, there is increasing evidence for additional functions of this histone modification. ATM-mediated H2AXS139ph plays a role during recombination of variable (V), diversity (D), and joining (J) segments, which is the basis for antigen receptor diversity during lymphocyte differentiation10. H2AXS139ph is important during angiogenesis since hypoxia-driven neovascularization is significantly reduced in endothelial-specific H2AX-deficient mice, and hypoxia induces H2AXS139ph in primary cultures of human endothelial cells9. Another study demonstrated that high basal levels of H2AXS139ph are required for stem cell self-renewal11. Our findings support a role of H2AXS139ph during Hmga2-induced gene activation. Accordingly, it has been reported that promoters of active (or poised) genes are either nucleosome-free or marked with unstable nucleosomes that contain specific histone variants such as H3.3 and H2A.Z28. Although H2AX does not affect nucleosome conformation, it has a destabilizing effect that is enhanced by its phosphorylation and results in an impaired linker histone H1 binding29. HMGA proteins were shown to dynamically compete with H1 for binding to the linker DNA, thereby loosening the chromatin30,31. In addition, displacement of H1 during the initial steps of gene activation has been reported32. Furthermore, the histone chaperone complex FACT (facilitates chromatin transcription) catalyzes integration of H2AX into nucleosomes33, induces disassembly of promoter-associated nucleosomes, and enhances transcriptional activation34. Interestingly, yeast FACT is a heterodimeric protein complex of Spt16 and Pob3 that interacts and functionally cooperates with the HMG1-like protein Nhp635. In summary, several lines of evidence and our work together support a role of HMG proteins and the chromatin modifications promoted by them during transcription initiation.
HMGA2 is a developmentally regulated protein, which is highly expressed in embryonic stem (ES) cells, almost absent in differentiated adult cells, but re-expressed in cancer3. EMT driven by TGFβ1 signaling occurs normally during development, and is also a major factor in the progression and metastasis of many cancer types. Hence, it would be interesting to study whether the mechanism of transcription regulation suggested here may contribute to ES cell differentiation or cancer development. This line of ideas gains further relevance within the context of embryonic lung development, where TGFβ1 and WNT signaling exert potential antagonizing effects. Here, HMGA2 has been shown to mediate TGF1 signaling5, and antagonizes canonical WNT signaling via Gata6 and Fzd216.
We propose a mechanism of transcriptional activation that requires HMGA2- and ATM-mediated H2AXS139ph. We show direct interaction between HMGA2 and ATM. In contrast to previous reports36, we did not detect interaction between HMGA2 and the other members of the superfamily of phosphatidylinositol 3-kinase-related kinases, ATR, and PRKDC. In addition, ATM has been related to chromatin changes that silence transcription in cis to DSBs37. These discrepancies may be explained by differences between the experimental systems used. On the other hand, there are also reports that link transcription activation to DSBs38,39,40, supporting the mechanism of transcriptional activation proposed here.
HMGA2, ATM, and H2AX have been related to biological events in which DNA-dependent processes such as repair, replication, recombination, and transcription are involved. Although packaging of the genome into chromatin constitutes a barrier for all these processes, the chromatin structure offers the possibility for a controlled implementation of all these processes. However, controlled chromatin disassembly might involve intermediates with SSB or even DSB that may compromise the integrity of DNA. Thus, there must be mechanisms that ensure the integrity of the genome during the opening of the chromatin. This perspective might explain the fact that proteins and histone modifications that were initially characterized during DNA repair also play a role during transcription, recombination, and replication. In line with this hypothesis, a common characteristic between HMGA2 transgenic41, ATM−/−42, and H2AX−/−43 mice is genomic instability. In addition, DNA repair-linked histone modifications, H3K56ac and H2Bub144, have been related to transcription initiation45. Remarkably, HMGA2 has been shown to have a lyase activity that cleaves DNA containing apurinic/apyrimidinic sites, thereby inducing base excision repair46. It will be the scope of future studies to investigate how the lyase activities of HMGA proteins affect the chromatin modifications and transcriptional activation reported here.
Materials and Methods
Cell culture
Mouse lung epithelial cells (MLE-12, ATCC CRL-2110), human embryonic kidney cells (HEK293T, ATCC CRL-11268), and NMuMG cells (ATCC CRL-1636) were obtained from the American Type Culture Collection. All cells were cultured following the supplier's instructions. Hmga2 stable knockdown MLE-12 cell line, FLAG-H2AX WT and S139A mutant stable HEK293T cell lines were generated; for detailed information, see Supplementary information, Data S1. Cells were transfected with plasmid DNA and/or siRNA using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions, and harvested 48 h later for further analysis. pcDNA3.1(A)-Hmga2-myc/his mammalian expression vector16 was used for Hmga2 overexpression in cells. siRNA control (siCtrl; D-001210-01-05; Dharmacon), siATM (SIHK0133; Sigma), siHMGA2 (M-013495-02; Dharmacon), and siHmga216 were transfected at 20 nM final concentration. ATM kinase inhibition was performed with 10 μM KU-55933 (118500-2MG; Calbiochem). HEK293T cells were treated with UV-C (80 J/m2) and H2O2 (1 mM), and harvested 6 h later for further analysis.
Dual-Luciferase reporter assays (Promega) were performed in HEK293T cells using pGL4-Gata6 promoter luciferase reporter plasmid16. For detailed information, see Supplementary information, Data S1.
Primary MEF were obtained from Hmga2+/+ and Hmga2−/− embryos at E15.5 and cultured as described47. MEF and MLE-12 cells were treated with 10 ng/ml and NMuMG cells with 5 ng/ml human recombinant TGFβ1 (240-B, R&D Systems). MLE-12 cells were treated with 1 mM L-mimosine (M0253, Sigma).
RNA isolation, reverse transcription, quantitative PCR, and affymetrix microarray data analysis
Total RNA was isolated with RNeasy® plus mini kit (Qiagen) and quantified using a nanodrop (Thermo Scientific, Germany). Synthesis of cDNA was performed using total RNA and the High Capacity cDNA Reverse Transcriptase kit (Applied Biosystem). Quantitative real-time PCR reactions were performed using SYBR® Green on the Step One plus Real-time PCR system (Applied Biosystem). Housekeeping genes Tuba1a and Gapdh were used to normalize gene expression48. Primer pairs used for gene expression analysis and Affymetrix microarray data analysis from WT and Hmga2−/− mouse embryonic lung at E18.5 (GEO: GSE55340) are described in Supplementary information, Data S1.
Chromatin immunoprecipitation (ChIP), and sequential ChIP (ChIP-reChIP) assays
ChIP analysis was performed as described16 with slight modifications. Briefly, cells were cross-linked by 1% formaldehyde for 10 min, lysed, and sonicated with Diagenode Biorupter to an average DNA length of 500-600 bp. After centrifugation, the soluble chromatin was immunoprecipitated using antibodies specific against phospho-(Ser/Thr) ATM substrate (Cell Signaling), H2AX (Millipore), H2AXS139ph (Ser139, Clone JBW301, Millipore), HMGA2 (Santa Cruz), RNA Pol II carboxy-terminal domain (4H8, Abcam), ATM (Santa Cruz), pSMAD3 (phospho S423+S425; Abcam), and IgG as a control (Santa Cruz). Precipitated chromatin complexes were removed from the beads through a 30 min incubation with 50 μl of 1% SDS, 0.1 M NaHCO3, vortexing each 5 min.
ChIP-reChIP experiment was performed49 with slight modifications using antibodies specific against phospho-(Ser/Thr) ATM substrate (Cell Signaling), H2AX (Millipore). Briefly, in re-ChIP experiments, complexes were eluted by incubation for 30 min at 37 °C in 25 μl 10 mM DTT. After centrifugation, the supernatant was diluted 20 times with re-ChIP buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, pH 8.1) and subjected again to the ChIP procedure.
Reverse cross-linked immunoprecipitated chromatin was subjected to qPCR. Primer pairs used for gene promoter analysis are described in Supplementary information, Data S1.
ChIP-seq
Single-end sequencing was performed on an Illumina HiSeq2500 machine at the Max Planck-Genome-Centre Cologne. ChIP-seq reads were mapped to mouse reference genome mm10 (UCSC assembly) using Bowtie2 with the default settings50. Peaks were called using MACS1.4 (P < 0.01) with a shift size of 30 bp51. ChIP-seq peak annotation and data visualization by preparing custom tracks on the UCSC genome browser was performed using HOMER. Distribution of the peaks in the genome regions (e.g., promoter, intron, intergenic, exon, 3′ UTR, and TTS) were done using KNIME SDK version 2.10.452. We used the Row filter and GroupBy functions to obtain the annotated peaks designed to the genome region. Enrichment of H2AXS139ph and ATM to the binding site of HMGA2 in the WT MEF was done using Homer annotatePeaks.pl by normalizing the tag density to 3 × 107 with a window size of 2 000 bp from the center of the peaks. To analyze the effect of HMGA2 in the binding of H2AXS139ph and ATM in the promoter region genome wide, we compared the Hmga2 WT with Hmga2−/− using ngs.plot53 and normalized the samples to their respective input.
Mass spectrometry: sample preparation, methods, and data analysis
MLE-12 cells nuclear protein lysates from control and HMGA2 immunoprecipitation were run on NuPAGE 4%-12% BIS-TRIS gel (Invitrogen, Carlsbad, USA). Gels were stained with colloidal blue (Invitrogen), and evenly sized gel pieces were excised and processed for mass spectrometry. The gel pieces were subjected to in gel reduction and alkylation, followed by trypsin digestion as described previously54.
Reverse phase nano-LC-MS/MS was performed by using an Agilent 1200 nanoflow LC system (Agilent Technologies, Santa Clara, CA, USA) using a cooled thermostated 96-well autosampler. The LC system was coupled with LTQ-Orbitrap instrument (Thermo Fisher Scientific) equipped with a nanoelectrospray source (Proxeon, Denmark). Chromatographic separation of peptides was performed in a 10-cm long and 75-μm inner diameter capillary needle. The column was custom-made with methanol slurry of reverse-phase ReproSil-Pur C18-AQ 3 μm resin (Dr Maisch GmbH). The tryptic peptide mixtures were autosampled at a flow rate of 0.5 μl/min and then eluted with a linear gradient at a flow rate 0.25 μl/min. The mass spectrometers were operated in the data-dependent mode to automatically measure MS and MS/MS spectra. LTQ-FT full scan MS spectra (from m/z 350 to 1 750) were acquired with a resolution of r = 60 000 at m/z 400. The five most intense ions were sequentially isolated and fragmented in the linear ion trap using collision-induced dissociation with collision energy of 35%. Further mass spectrometric parameters: spray voltage of 2.4 kV, no sheath gas flow, and the temperature of the heated capillary was 200 °C55.
For data analysis, we used the MaxQuant software tool56, Version 1.2.0.8. The measured raw data were processed and quantitated as described in57.
Animal experiments
C57BL/6 and Hmga2+/− mice (stock # 002644, Jackson Laboratories)13 were obtained from Charles River Laboratories at 5-6 weeks of age. Hmga2 overexpression mice were kind gift from Prof. Monica Bessler22. Animals were housed and bred under controlled temperature and lighting (12/12-h light/dark cycle), fed with commercial animal feed and water. Timed pregnant mice were killed at E18.5 and embryonic lungs were harvested for ChIP, western blot, and expression analysis. Mouse work was performed in compliance with the German Law for Welfare of Laboratory Animals, which complies with international regulations. The permission to perform the experiments presented in this study was obtained from the Regional Council (Regierungspräsidium in Darmstadt, Germany; IVMr46-53r30.03.MPP04.12.02 and IVMr46-53r30.03.MPP06.12.01).
Statistical analysis
Samples were analyzed at least in triplicates and experiments were performed three times. Statistical analysis was performed using Excel Solver. All data are represented as mean ± standard error (mean ± sem). One-way analysis of variance (ANOVA) was used to determine the levels of difference between the groups and P values for significance. P values after one-way ANOVA, *P ≤ 0.05; **P < 0.01, and ***P < 0.001. For the ChIP-seq data, P values were calculated on R statistical platform using Fisher's exact test. P < 0.0001 is considered extremely significant.
The Affymetrix microarray-based transcriptome data have been deposited in NCBI's Gene Expression Omnibus58 and is accessible through GEO Series with accession number GSE55340. The mass spectrometry-based HMGA2 interactome data have been deposited in EMBL-EBI with the accession number EBI-9985175 and the ChIP-seq data have been deposited in NCBI with the accession number GSE63861.
For more details of experimental procedures refer to Supplementary information available with this article online.
References
Bustin M, Catez F, Lim JH . The dynamics of histone H1 function in chromatin. Mol Cell 2005; 17:617–620.
Catez F, Hock R . Binding and interplay of HMG proteins on chromatin: lessons from live cell imaging. Biochim Biophys Acta 2010; 1799:15–27.
Ozturk N, Singh I, Mehta A, Braun T, Barreto G . HMGA proteins as modulators of chromatin structure during transcriptional activation. Front Cell Dev Biol 2014; 2:5.
Reeves R . Nuclear functions of the HMG proteins. Biochim Biophys Acta 2010; 1799:3–14.
Thuault S, Valcourt U, Petersen M, Manfioletti G, Heldin CH, Moustakas A . Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. J Cell Biol 2006; 174:175–183.
Lavin MF . Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol 2008; 9:759–769.
Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM . DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998; 273:5858–5868.
Redon CE, Nakamura AJ, Martin OA, Parekh PR, Weyemi US, Bonner WM . Recent developments in the use of gamma-H2AX as a quantitative DNA double-strand break biomarker. Aging 2011; 3:168–174.
Economopoulou M, Langer HF, Celeste A, et al. Histone H2AX is integral to hypoxia-driven neovascularization. Nat Med 2009; 15:553–558.
Steinel NC, Lee BS, Tubbs AT, et al. The ataxia telangiectasia mutated kinase controls Igkappa allelic exclusion by inhibiting secondary Vkappa-to-Jkappa rearrangements. J Exp Med 2013; 210:233–239.
Turinetto V, Orlando L, Sanchez-Ripoll Y, et al. High basal gammaH2AX levels sustain self-renewal of mouse embryonic and induced pluripotent stem cells. Stem Cells 2012; 30:1414–1423.
McManus KJ, Hendzel MJ . ATM-dependent DNA damage-independent mitotic phosphorylation of H2AX in normally growing mammalian cells. Mol Biol Cell 2005; 16:5013–5025.
Xiang X, Benson KF, Chada K . Mini-mouse: disruption of the pygmy locus in a transgenic insertional mutant. Science 1990; 247:967–969.
Bakkenist CJ, Kastan MB . DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 2003; 421:499–506.
Mukherjee P, Winter SL, Alexandrow MG . Cell cycle arrest by transforming growth factor beta1 near G1/S is mediated by acute abrogation of prereplication complex activation involving an Rb-MCM interaction. Mol Cell Biol 2010; 30:845–856.
Singh I, Mehta A, Contreras A, et al. Hmga2 is required for canonical WNT signaling during lung development. BMC Biol 2014; 12:21.
Zhou X, Benson KF, Ashar HR, Chada K . Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 1995; 376:771–774.
Gagou ME, Zuazua-Villar P, Meuth M . Enhanced H2AX phosphorylation, DNA replication fork arrest, and cell death in the absence of Chk1. Mol Biol Cell 2010; 21:739–752.
Bonner WM, Redon CE, Dickey JS, et al. GammaH2AX and cancer. Nat Rev Cancer 2008; 8:957–967.
Szuts D, Krude T . Cell cycle arrest at the initiation step of human chromosomal DNA replication causes DNA damage. J Cell Sci 2004; 117:4897–4908.
Chung LC, Tsui KH, Feng TH, Lee SL, Chang PL, Juang HH . L-Mimosine blocks cell proliferation via upregulation of B-cell translocation gene 2 and N-myc downstream regulated gene 1 in prostate carcinoma cells. Am J Physiol Cell Physiol 2012; 302:C676–685.
Ikeda K, Mason PJ, Bessler M . 3′UTR-truncated Hmga2 cDNA causes MPN-like hematopoiesis by conferring a clonal growth advantage at the level of HSC in mice. Blood 2011; 117:5860–5869.
Chen J, Herrup K . Glutamine acts as a neuroprotectant against DNA damage, beta-amyloid and H2O2-induced stress. PloS One 2012; 7:e33177.
Knobel PA, Belotserkovskaya R, Galanty Y, Schmidt CK, Jackson SP, Stracker TH . USP28 is recruited to sites of DNA damage by the tandem BRCT domains of 53BP1 but plays a minor role in double-strand break metabolism. Mol Cell Biol 2014; 34:2062–2074.
Smith JJ, Cole ES, Romero DP . Transcriptional control of RAD51 expression in the ciliate Tetrahymena thermophila. Nucleic Acids Res 2004; 32:4313–4321.
Youn CK, Song PI, Kim MH, et al. Human 8-oxoguanine DNA glycosylase suppresses the oxidative stress induced apoptosis through a p53-mediated signaling pathway in human fibroblasts. Mol Cancer Res 2007; 5:1083–1098.
Ma Z, Yao G, Zhou B, Fan Y, Gao S, Feng X . The Chk1 inhibitor AZD7762 sensitises p53 mutant breast cancer cells to radiation in vitro and in vivo. Mol Med Rep 2012; 6:897–903.
Jin C, Zang C, Wei G, et al. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat Genet 2009; 41:941–945.
Li A, Yu Y, Lee SC, Ishibashi T, Lees-Miller SP, Ausio J . Phosphorylation of histone H2A.X by DNA-dependent protein kinase is not affected by core histone acetylation, but it alters nucleosome stability and histone H1 binding. J Biol Chem 2010; 285:17778–17788.
Kishi Y, Fujii Y, Hirabayashi Y, Gotoh Y . HMGA regulates the global chromatin state and neurogenic potential in neocortical precursor cells. Nat Neurosci 2012; 15:1127–1133.
Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M . Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol 2004; 24:4321–4328.
Vicent GP, Nacht AS, Font-Mateu J, et al. Four enzymes cooperate to displace histone H1 during the first minute of hormonal gene activation. Genes Dev 2011; 25:845–862.
Heo K, Kim H, Choi SH, et al. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol Cell 2008; 30:86–97.
Ransom M, Williams SK, Dechassa ML, et al. FACT and the proteasome promote promoter chromatin disassembly and transcriptional initiation. J Biol Chem 2009; 284:23461–23471.
Formosa T, Eriksson P, Wittmeyer J, Ginn J, Yu Y, Stillman DJ . Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J 2001; 20:3506–3517.
Natarajan S, Hombach-Klonisch S, Droge P, Klonisch T . HMGA2 inhibits apoptosis through interaction with ATR-CHK1 signaling complex in human cancer cells. Neoplasia 2013; 15:263–280.
Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA . ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 2010; 141:970–981.
Ju BG, Lunyak VV, Perissi V, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science 2006; 312:1798–1802.
Lin C, Yang L, Tanasa B, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell 2009; 139:1069–1083.
Hamperl S, Cimprich KA . The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Rep 2014; 19:84–94.
Li AY, Boo LM, Wang SY, et al. Suppression of nonhomologous end joining repair by overexpression of HMGA2. Cancer Res 2009; 69:5699–5706.
Barlow C, Hirotsune S, Paylor R, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell 1996; 86:159–171.
Celeste A, Petersen S, Romanienko PJ, et al. Genomic instability in mice lacking histone H2AX. Science 2002; 296:922–927.
Kari V, Shchebet A, Neumann H, Johnsen SA . The H2B ubiquitin ligase RNF40 cooperates with SUPT16H to induce dynamic changes in chromatin structure during DNA double-strand break repair. Cell Cycle 2011; 10:3495–3504.
Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M . Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol 2008; 10:483–488.
Summer H, Li O, Bao Q, et al. HMGA2 exhibits dRP/AP site cleavage activity and protects cancer cells from DNA-damage-induced cytotoxicity during chemotherapy. Nucleic Acids Res 2009; 37:4371–4384.
Winslow MM, Dayton TL, Verhaak RG, et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature 2011; 473:101–104.
Mehta A, Dobersch S, Dammann RH, et al. Validation of Tuba1a as appropriate internal control for normalization of gene expression analysis during mouse lung development. Int J Mol Sci 2015; 16:4492–4511.
Metivier R, Penot G, Hubner MR, et al. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 2003; 115:751–763.
Langmead B, Salzberg SL . Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9:357–359.
Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008; 9:R137.
Jagla B, Wiswedel B, Coppee JY . Extending KNIME for next-generation sequencing data analysis. Bioinformatics 2011; 27:2907–2909.
Shen L, Shao N, Liu X, Nestler E . ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 2014; 15:284.
Shevchenko A, Jensen ON, Podtelejnikov AV, et al. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci USA 1996; 93:14440–14445.
Mukhametshina RT, Ruhs A, Singh I, et al. Quantitative proteome analysis of alveolar type-II cells reveals a connection of integrin receptor subunits beta 2/6 and WNT Signaling. J Proteome Res 2013; 12:5598–5608.
Cox J, Mann M . MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008; 26:1367–1372.
Cox J, Matic I, Hilger M, et al. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat Protoc 2009; 4:698–705.
Edgar R, Domrachev M, Lash AE . Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002; 30:207–210.
Acknowledgements
We thank Bender R and Goth K for technical support; Bessler M, Kwon J for reagents; and Dimmeler S, Renkawitz R and Schaprian A for helpful discussions. Barreto G is funded by the “LOEWE-Initiative der Landesförderung” (III L4–18/15.004 2009) and the DFG grant BA 4036/1-1. R. Mostoslavsky is supported by NIH grants R01GM093072-01. Cosentino C receives a Fellowship from the Fondazione Umberto Veronesi. Bellusci S is supported by HL086322, HL074832 and the ECCPS. This work was done according to the program of competitive growth of the Kazan Federal University and the Russian Government.
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
Quality analysis of ChIP-seq data for HMGA2, H2AXS139ph and ATM in Hmga2 +/+ and −/− MEF. (PDF 105 kb)
Supplementary information, Figure S2
(A) Visualization of selected genes using UCSC genome browser shows occupancy of HMGA2 (blue), H2AXS139ph (green) and ATM (maroon) in Hmga2 +/+ and −/− MEF. (PDF 205 kb)
Supplementary information, Figure S3
(A) KU-55933 (ATMi) inhibits kinase activity of ATM. (PDF 83 kb)
Supplementary information, Figure S4
Hmga2-KO reduced whereas Hmga2-GOF increased the levels of H2AXS139ph on the Gata6 promoter. (PDF 95 kb)
Supplementary information, Figure S5
(A) Confirmation of result presented in Figure 5A: Interaction of ATM with H2AX is independent of H2AX phosphorylation site S139. (PDF 82 kb)
Supplementary information, Data S1
Materials and Methods (PDF 58 kb)
Rights and permissions
About this article
Cite this article
Singh, I., Ozturk, N., Cordero, J. et al. High mobility group protein-mediated transcription requires DNA damage marker γ-H2AX. Cell Res 25, 837–850 (2015). https://doi.org/10.1038/cr.2015.67
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2015.67
Keywords
This article is cited by
-
HMGA2 directly mediates chromatin condensation in association with neuronal fate regulation
Nature Communications (2023)
-
Autophagy inhibitor-loaded mesoporous AgNPs@SiO2 nanoplatform for synergistically enhanced glioma radiotherapy
Science China Materials (2023)
-
Lymphoid-specific helicase in epigenetics, DNA repair and cancer
British Journal of Cancer (2022)
-
Positioning of nucleosomes containing γ-H2AX precedes active DNA demethylation and transcription initiation
Nature Communications (2021)
-
Knockdown of HMGA2 regulates the level of autophagy via interactions between MSI2 and Beclin1 to inhibit NF1-associated malignant peripheral nerve sheath tumour growth
Journal of Experimental & Clinical Cancer Research (2019)