Abstract
Recent studies have boosted our understanding of long noncoding RNAs (lncRNAs) in numerous biological processes, but few have examined their roles in somatic cell reprogramming. Through expression profiling and functional screening, we have identified that the large intergenic noncoding RNA p21 (lincRNA-p21) impairs reprogramming. Notably, lincRNA-p21 is induced by p53 but does not promote apoptosis or cell senescence in reprogramming. Instead, lincRNA-p21 associates with the H3K9 methyltransferase SETDB1 and the maintenance DNA methyltransferase DNMT1, which is facilitated by the RNA-binding protein HNRNPK. Consequently, lincRNA-p21 prevents reprogramming by sustaining H3K9me3 and/or CpG methylation at pluripotency gene promoters. Our results provide insight into the role of lncRNAs in reprogramming and establish a novel link between p53 and heterochromatin regulation.
Similar content being viewed by others
Introduction
Induced pluripotent stem cells (iPSCs) have major implications for regenerative medicine, in vitro disease modeling and toxicology screening1. Yet, to fulfill these expectations, it is necessary to understand how the reprogramming machinery erases and rewrites the somatic epigenetic code. This is important because, among other reasons, reprogramming is prone to errors that could hamper biomedical applications of the derived cells2.
Reprogramming by exogenous factors is nowadays considered as a multistep process in which cells must overcome a series of roadblocks to ultimately acquire pluripotency3,4, though under certain conditions it can also be deterministic5,6,7. Two fundamental reprogramming roadblocks are the activation of a p53-mediated response that promotes apoptosis and cell senescence, and the inefficient removal of preexisting somatic-like epigenetic marks. Accordingly, depletion of p538 or the H3K9 methyltransferases Setdb1, Suv39h1 and Suv39h29,10,11, and inhibition of the maintenance DNA methyltransferase DNMT112 improve reprogramming efficiency.
Noncoding RNAs have important roles in establishing or removing different reprogramming roadblocks. However, despite the extensive characterization of microRNA (miRNA) networks in reprogramming13, only 2 long noncoding RNAs (lncRNAs) are known to modulate this process. LncRNAs are noncoding RNA species longer than 200 nucleotides and act as decoys, guides or scaffolds14. LincRNA-RoR was the first lncRNA implicated in reprogramming15 and partly functions as a 'sponge' that titrates down miRNAs targeting pluripotency regulators (e.g., miR-145)16. Conversely, lincRNA TUNA forms a complex with several RNA-binding proteins at the promoters of Nanog, Sox2 and Fgf4, where it facilitates the deposition of the active histone mark H3K4me317.
In this report, we identify that the p53-induced large intergenic noncoding RNA p21 (lincRNA-p21; also termed Trp53cor1)18,19,20 acts as a barrier for reprogramming by sustaining heterochromatic state at pluripotency gene promoters.
Results
Functional screening reveals roles of lincRNAs in the preiPSC to iPSC conversion
Somatic cells undergoing reprogramming can be trapped in a partially reprogrammed state termed the preiPSC state21. These cells have overcome the senescence barrier and downregulated multiple somatic markers, but fail to fully activate the pluripotency network. Importantly, preiPSCs are stable and can be readily converted to fully reprogrammed iPSCs through a variety of approaches10,21. Because reprogramming cells consist of heterogeneous populations progressing at different speeds3, preiPSCs provide a relatively well-defined model for dissecting the chain of events eventually leading to the full acquisition of pluripotency10,11. Using preiPSCs, we thus sought to identify lncRNAs that control the late stage of mouse somatic cell reprogramming.
We selected 51 lincRNAs previously reported to regulate embryonic stem cell (ESC) pluripotency (26) or differentiation (25)22 and 16 p53-regulated lincRNAs18 (Figure 1A). The rationale for choosing the latter group is that p53 is an important regulator of ESC differentiation and a major barrier for reprogramming8. We performed quantitative PCR (qPCR) analysis of these 67 lincRNAs in mouse embryonic fibroblasts (MEFs), 4 independent preiPSC clones (generated from Oct4-GFP transgenic MEFs21 using Oct4, Sox2, Klf4 and c-Myc; OSKM), iPSCs and ESCs. The analysis showed distinct patterns of lincRNA expression in the 4 cell types that were grouped into 7 classes (Figure 1B, 1C and Supplementary information, Table S1A). Classes I and II were the largest and contained 17 and 26 lincRNAs, respectively, while class VI included only 1 lincRNA (p53-regulated) (Supplementary information, Table S1A). In contrast to classes I-VI, class VII lincRNAs did not show a consistent pattern among the different cell types. Of note, pluripotency- and differentiation-related lincRNAs were distributed evenly among classes I-IV and VII, but only 1 differentiation-related lincRNA was included in class V. Conversely, p53-regulated lincRNAs were mostly in classes II (8) and V (comprising 4 out of 5 constituents). We also performed qPCR analysis for all 67 lincRNAs in MEFs reprogrammed with OSKM (Supplementary information, Figure S1 and Table S1B), and observed that 58 out of the 67 lincRNAs increase by at least 2-fold during reprogramming (Supplementary information, Table S1B).
Functional screening of lincRNAs in the preiPSC to iPSC conversion. (A) Schematic view of the screening. (B) Heatmap of 67 lincRNAs quantified by qPCR in the 4 indicated cell types. The expression level of individual lincRNAs was normalized to MEFs. Colors represent fold change above (red) or below (blue) the values in MEFs. (C) Box plot analysis of lincRNA expression levels in the indicated cell types. I-VI indicates distinct expression patterns as shown in B. PSCs = iPSCs and ESCs. (D) Upper panel, knockdown efficiency of shRNAs for the indicated lincRNAs in a selected preiPSC clone. Data are presented as mean ± SD of 3 independent experiments with triplicate items (also in the lower panel). shRNA targeting firefly luciferase was used as control (shCtrl; also hereafter in similar experiments). Lower panel, relative number of GFP+ colonies arising from the same preiPSC clone after lincRNA shRNA knockdown and treatment with Vc compared to control shRNA. *P < 0.05, **P < 0.01, ***P < 0.001 by the Student's t-test (also hereafter). (E) Representative images of GFP+ colonies produced as in D. Bright-field (phase) is also shown. Scale bar, 100 μm. Supportive data are included in Supplementary information, Figure S1 and Table S1.
We then selected 15 lincRNAs from classes I-VI for knockdown and functional analysis in a validated preiPSC to iPSC conversion model10. We generated 2 individual short hairpin RNA (shRNA) viral vectors for each of the 15 lincRNAs, and identified at least 1 effective shRNA (> 50% depletion) for 11 out of the 15 lincRNAs (Figure 1D, upper panel). Activation of the Oct4-GFP reporter was employed as readout for detecting the full acquisition of pluripotency. After infection with the shRNAs, the preiPSCs were passaged onto feeders and treated with vitamin C (Vc) for 8 days before GFP+ colony counting10. Notably, knockdown of 3 lincRNAs (-1463, -1526, and -p21) with the 2 independent shRNAs showed enhanced GFP+ colony formation compared to the control, while lincRNA-1307 knockdown had the opposite effects (Figure 1D, lower panel and 1E). In summary, our screening successfully identified 4 lincRNAs that modulate the preiPSC to iPSC conversion.
p53-induced lincRNA-p21 impairs reprogramming independent of apoptosis and cell proliferation
Among the 4 lincRNAs identified above, we considered lincRNA-p21 particularly interesting because of its expression profile (class V: higher in preiPSCs compared to iPSCs/ESCs and MEFs) and p53-dependent regulation. LincRNA-p21 mediates the apoptotic effects of p53 upon DNA damage18, although we did not detect any change in apoptosis or cell proliferation during the conversion of preiPSCs to iPSCs when lincRNA-p21 was knocked down (data not shown). Of note, p53 protein levels were higher in preiPSCs compared with MEFs and iPSCs/ESCs (Figure 2A). This was not due to increased p53 mRNA expression (Supplementary information, Figure S2A), indicating that p53 is likely regulated at the post-transcriptional level in this setting. Intriguingly, p53 knockdown similarly enhanced the conversion of preiPSCs to iPSCs (Figure 2B) without influencing apoptosis or proliferation (Supplementary information, Figure S2B and S2C). These findings suggest that p53 and lincRNA-p21 act in the same pathway, independent of apoptosis or proliferation, to sustain the preiPSC state.
p53-induced lincRNA-p21 is a barrier for reprogramming. (A) Representative western blot for p53 in the 4 indicated cell types. ACTIN was the loading control. (B) Relative number of GFP+ colonies arising from 2 independent preiPSC clones cotransduced with control shRNA or 2 independent shRNAs for p53. Data are presented as mean ± SD of 3 independent experiments with triplicate items (also in D, I, J and K). (C) qPCR analysis for lincRNA-p21 in MEFs cotransduced with OSKM and either control shRNA or 2 independent shRNAs for p53. Data are presented as mean ± SD of 2 independent experiments with triplicate items (also in F). (D) Left panel, qPCR analysis for lincRNA-p21 in MEFs cotransduced with OSKM and either control shRNA or 2 independent shRNAs for lincRNA-p21 on day 6. Right panel, relative number of GFP+ colonies counted at day 15 (also in I, J and K) in similar reprogramming experiments. (E) Apoptosis in MEFs cotransduced with OSKM and either control shRNA or 2 independent shRNAs for lincRNA-p21. Samples were analyzed on day 6. shRNA for p53 was used as positive control (also in F). Data are presented as mean ± SD of a representative experiment with triplicate items. (F) Cell proliferation in MEFs cotransduced with OSKM and either control shRNA or 2 independent shRNAs for lincRNA-p21. (G) Representative immunofluorescence, phase contrast photographs and photographs of chimeric mice obtained with an iPSC clone produced with OSKM and shRNA for lincRNA-p21. Scale bar, 50 μm (left panels) and 200 μm (right panels). (H) Karyotype analysis for the same iPSC clone employed for chimeric mouse generation in G. (I) Relative number of GFP+ colonies in MEFs cotransduced with OSKM and lincRNA-p21. Empty vector was used as control (control; also hereafter in similar experiments). (J) Relative number of GFP+ colonies in MEFs cotransduced with OSKM and the indicated combinations of lincRNA-p21 and shRNAs. (K) Relative number of GFP+ colonies in MEFs co-transduced with OSKM and either control shRNA, shRNA for lincRNA-p21 or shRNA for lincRNA-p21 plus shRNA-resistant lincRNA-p21. Supportive data are included in Supplementary information, Figures S2 and S3.
We also detected that lincRNA-p21 expression increases significantly (from ∼20 to ∼300 copies per cell) during the reprogramming of MEFs with OSKM (Figure 2C and Supplementary information, Figure S2D and Table S1B). This induction was p53-dependent, as assessed by cotransduction of p53 shRNAs and OSKM into wild-type MEFs and the reprogramming of p53−/− tail tip fibroblasts with OSKM (Figure 2C and Supplementary information, Figure S2E). We then tested the effect of knocking down lincRNA-p21 whilst simultaneously reprogramming Oct4-GFP MEFs with OSKM. This led to a higher number of GFP+ colonies (∼4-6-fold) compared to the control without noticeably influencing apoptosis or cell proliferation, as opposed to the effect of p53 knockdown (Figure 2D-2F). Besides, we observed that lincRNA-p21 depletion does not exhibit synergy with p53 depletion in either cell proliferation or reprogramming efficiency (Supplementary information, Figure S3A and S3B). Of note, the resulting iPSC colonies produced with lincRNA-p21 knockdown were fully reprogrammed, as demonstrated by standard characterization procedures including the generation of chimeric mice (Figure 2G and Supplementary information, Figure S3C and S3D). Likewise, they had integrated the shRNA vectors into their genome (Supplementary information, Figure S3E) and displayed a normal karyotype (Figure 2H). Moreover, we observed that overexpressing lincRNA-p21 impairs OSKM reprogramming efficiency (Figure 2I) and diminishes the enhancing effect of p53 knockdown in a similar manner (Figure 2J). In addition, overexpressing a shRNA-resistant lincRNA-p21 with OSKM reduced the enhanced reprogramming efficiency achieved by lincRNA-p21 knockdown (Figure 2K), ruling out an off-target effect of the shRNA. Therefore, p53-induced lincRNA-p21 acts as a barrier for reprogramming independent of apoptosis and cell proliferation.
LincRNA-p21 associates with SETDB1 and DNMT1 in separate repressor complexes facilitated by HNRNPK
During the DNA damage response, lincRNA-p21 interacts with and directs the RNA-binding protein HNRNPK to specific genomic loci that are subsequently silenced by an unknown mechanism18. We thus speculated that lincRNA-p21 might interact with alternative partners in reprogramming cells to regulate a distinctive set of genes. Specifically, we envisaged that these partners would be chromatin modifiers with a repressive role in reprogramming. To explore this idea, we first performed cellular fractionation to analyze the subcellular localization of lincRNA-p21 during the reprogramming of MEFs with OSKM. LincRNA-p21 was predominantly nuclear (Figure 3A), consistent with a potential role in epigenetic regulation. Then, we employed RNA immunoprecipitation (RIP) with antibodies against a panel of chromatin modifiers detrimental for reprogramming9,10,11,12,23 using lysates from MEFs reprogrammed with OSKM on day 12. Notably, there was a substantial enrichment for lincRNA-p21 in DNMT1 RIP and a less substantial enrichment in SETDB1 RIP, but no enrichment in DNMT3A/B and SUV39H1 RIPs, relative to IgG control (Figure 3B). The enrichment for lincRNA-p21 in DNMT1 and SETDB1 RIPs was also present at an earlier phase of OSKM reprogramming (Supplementary information, Figure S4A). We also observed that both SETDB1 and DNMT1 increase during OSKM reprogramming (Supplementary information, Figure S4B).
LincRNA-p21 and HNRNPK associate with SETDB1 and DNMT1 in reprogramming. (A) Representative analysis of lincRNA-p21 distribution by cellular fractionation of MEFs reprogrammed with OSKM on day 6. U6 RNA and Gapdh mRNA served as controls for nuclear and cytoplasmic RNAs, respectively. (B) RIP analysis of DNMT1, DNMT3A, DNMT3B, SUV39H1 and SETDB1 in MEFs reprogrammed with OSKM on day 12. Data are presented as mean ± SD of 2 independent experiments with triplicate items. (C) Representative immunoprecipitation (IP) of SETDB1 (left) or DNMT1 (right) in MEFs reprogrammed with OSKM on day 12. The membranes were immunoblotted (IB) for HNRNPK, SETDB1 or DNMT1. Specific bands are marked with an arrow (also in D). (D) Representative IP of HNRNPK in MEFs reprogrammed with OSKM on day 12. The membranes were immunoblotted for HNRNPK, SETDB1 or DNMT1. (E) In vitro RIP analysis of SETDB1 using purified lincRNA-p21, SETDB1-Flag and HNRNPK-Flag at different concentrations. The extracted RNA was then analyzed by qPCR (upper) and semi-qPCR (lower). Data are presented as mean ± SD of 2 independent experiments with triplicate items (upper) and a representative experiment (lower). Supportive data are included in Supplementary information, Figure S4.
To confirm the above interactions, we performed RNA pull down with in vitro transcribed biotinylated lincRNA-p21 and lacZ control using nuclear extracts of MEFs reprogrammed with OSKM. Yet, despite a robust interaction with HNRNPK, we failed to detect SETDB1 or DNMT1 by western blotting (Supplementary information, Figure S4C). This led us to speculate that the interaction between lincRNA-p21, SETDB1 and DNMT1 is facilitated by another protein, potentially HNRNPK. To demonstrate this, we conducted immunoprecipitation of endogenous SETDB1 and DNMT1 in MEFs reprogrammed with OSKM on day 12, and observed that endogenous HNRNPK could be pulled down in both cases (Figure 3C). This interaction was confirmed by reverse immunoprecipitation using antibodies against HNRNPK (Figure 3D). However, we did not detect interaction between SETDB1 and DNMT1, indicating that each of them likely associates separately with HNRNPK (data not shown). We also performed an in vitro interaction assay with purified HNRNPK, SETDB1 and lincRNA-p21. The result showed that SETDB1 interacts with lincRNA-p21 in an HNRNPK-dependent manner (Figure 3E), though we cannot exclude the possibility that additional proteins present in the lysates facilitate the interaction. Besides, we observed decreased enrichment for lincRNA-p21 in SETDB1 or DNMT1 RIPs upon Hnrnpk knockdown in MEFs reprogrammed with OSKM on day 12 (Figure 4A and 4B). Yet, this was not associated with reduced SETDB1 or DNMT1 protein or mRNA expression (Supplementary information, Figure S4D and S4E), which reinforces the notion that HNRNPK facilitates the interaction between SETDB1/DNMT1 and lincRNA-p21 to impair reprogramming. Supporting this, we observed that Hnrnpk expression increases in reprogramming (Supplementary information, Figure S4F and S4G). Moreover, Hnrnpk knockdown using shRNAs enhanced reprogramming efficiency (∼5-6-fold) (Figure 4C) and its overexpression blocked reprogramming (Figure 4D). Likewise, the effect of Hnrnpk knockdown was not synergistic with the effect of lincRNA-p21 knockdown (Figure 4E), further arguing that Hnrnpk and lincRNA-p21 act in the same pathway. In addition, we noticed enhanced conversion of preiPSCs to iPSCs with Hnrnpk knockdown upon treatment with Vc (Figure 4F). Altogether, these experiments indicate that lincRNA-p21 and SETDB1 or DNMT1 associate in separate complexes in reprogramming cells, which are facilitated by HNRNPK.
HNRNPK regulates reprogramming. (A, B) RIP analysis of SETDB1 (A) or DNMT1 (B) in MEFs cotransduced with OSKM and either control shRNA or an shRNA for Hnrnpk on day 12. The concentration of lincRNA-p21 (measured by qPCR) in the different input samples (shRNA for Hnrnpk or control) employed for RIP was comparable. Data are presented as mean ± SD of 3 independent experiments with triplicate items (also in C, D, E and F). (C) Relative number of GFP+ colonies counted on day 15 (also in D and E) in MEFs cotransduced with OSKM and either control shRNA or 2 independent shRNAs for Hnrnpk. (D) Relative number of GFP+ colonies in MEFs reprogrammed with OSKM and overexpressed HNRNPK. (E) Relative number of GFP+ colonies in MEFs cotransduced with OSKM and the indicated shRNAs. NS = not significant. (F) Relative number of GFP+ colonies arising from 2 independent preiPSC clones transduced with control shRNA or 2 independent shRNAs for Hnrnpk. Supportive data are included in Supplementary information, Figure S4.
LincRNA-p21 sustains H3K9me3 and CpG methylation at pluripotency genes
To identify the genes targeted by lincRNA-p21 during reprogramming and better understand the global interaction with HNRNPK, we performed RNA-sequencing (RNA-seq) of MEFs reprogrammed with OSKM with or without lincRNA-p21 or Hnrnpk knockdown (2 independent shRNAs for each target and 2 shRNA control) at 3 different time points. Notably, most of the genes differentially regulated by lincRNA-p21 knockdown at each time point (compared with the control) were among those genes differentially regulated by Hnrnpk knockdown (Figure 5A, 5B and Supplementary information, Figure S5A and Table S2). This result indicated that lincRNA-p21 mainly exerts its role in reprogramming through HNRNPK. KEGG pathway analysis of genes upregulated by lincRNA-p21 or Hnrnpk knockdown included the p53 signaling pathway (Supplementary information, Figure S5B). In addition, principal component analysis of the RNA-seq time course similarly confirmed the direction of global gene expression, with a dramatic divergence between the control and the lincRNA-p21, as well as Hnrnpk knockdown (Figure 5C).
The function of LincRNA-p21 in reprogramming is mainly mediated by HNRNPK. (A) Venn diagrams show the overlap of genes significantly differentially upregulated (> 1.5-fold) by lincRNA-p21 or Hnrnpk knockdown in MEFs cotransduced with OSKM. (B) Heatmap shows all genes significantly differentially regulated (q-value < 0.05) by lincRNA-p21 and Hnrnpk knockdown. Colors represent fold-change levels above (red) or below (blue) the control. Selected genes are indicated. (C) Principal component analysis of the variance between samples. D = day. (D) qPCR analysis for the indicated pluripotency genes in MEFs cotransduced with OSKM and either control shRNA or 2 independent shRNAs for lincRNA-p21 on day 12. Data are presented as mean ± SD of 3 independent experiments with triplicate items. Supportive data are included in Supplementary information, Figures S5, S6 and Table S2.
Importantly, mesenchymal and epithelial genes showed no difference upon lincRNA-p21 or Hnrnpk knockdown in the RNA-seq time course compared with the control (Supplementary information, Table S2), which was validated by qPCR (Supplementary information, Figure S6A). We did not observe any change in the cell cycle repressor p21 in the RNA-seq time course either (Supplementary information, Table S2), which supports our data that lincRNA-p21 knockdown does not increase cell proliferation in the preiPSC to iPSC conversion or in reprogramming. However, many pluripotency regulators such as Esrrb, Lin28a, Utf1, Nr5a2, Sall4, Lefty2 and Nanog were significantly upregulated by lincRNA-p21 or Hnrnpk knockdown (Figure 5B and Supplementary information, Table S2). We performed qPCR analysis to confirm that the pluripotency network is severely affected by lincRNA-p21 knockdown (Figure 5D). Interestingly, Lin28 had been previously described as a target for lincRNA-p21 in MEFs subjected to DNA damage18. As a major function of LIN28 is to block the maturation of let-7 miRNAs, we measured mature let-7 miRNAs by qPCR and observed that they were downregulated (Supplementary information, Figure S6B).
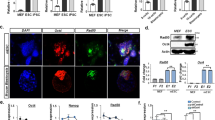
Next, we employed chromatin immunoprecipitation (ChIP) to assess the binding of HNRNPK to the promoters of a panel of pluripotency genes in MEFs reprogrammed with OSKM on day 12. There was a significant enrichment for HNRNPK compared to the IgG control in most of the selected targets (Supplementary information, Figure S7A), and this was reduced in MEFs cotransduced with shRNA for lincRNA-p21 compared to control shRNA (Figure 6A). A similar pattern was observed when we analyzed the binding of SETDB1 and DNMT1 to the promoters of the same panel of genes (Figure 6B and 6C), though DNMT1 binding to Nanog was more enriched compared to SETDB1. Likewise, lincRNA-p21 knockdown reduced H3K9me3 enrichment in the same genes bound by HNRNPK (except for Nanog) compared to the shRNA control (Figure 6D). In addition, Setdb1 and lincRNA-p21 knockdown produced a similar reduction in H3K9me3 at target promoters and their combination was not synergistic (Figure 6D). On the other hand, analysis of CpG methylation by bisulfite sequencing demonstrated that lincRNA-p21 and Dnmt1 knockdown accelerate the demethylation of the Nanog promoter compared to the control and their combination is not synergistic (Figure 6E). However, lincRNA-p21 knockdown had little effect on CpG methylation of the Oct4 promoter (Supplementary information, Figure S7B), in agreement with the observation that DNMT1 binding to the Oct4 promoter was not affected by lincRNA-p21 knockdown (Figure 6C). We could not detect significant methylation in the Lin28a promoter region that we amplified (data not shown).
LincRNA-p21 regulates H3K9me3 and CpG methylation at pluripotency genes. (A-C) ChIP analysis of MEFs cotransduced with OSKM and control shRNA or shRNA for lincRNA-p21 on day 12. Binding of HNRNPK (A), SETDB1 (B) and DNMT1 (C) at the promoters of the indicated pluripotency genes was quantified by qPCR and shown as relative enrichment compared with IgG. Gapdh promoter served as control. Data are presented as mean ± SD of 3 (A) or 2 (B, C and D) independent experiments with triplicate items. (D) ChIP analysis of MEFs cotransduced with OSKM and the indicated combinations on day 12. H3K9me3 enrichment at the promoters of the indicated pluripotency genes were quantified by qPCR and then normalized to total histone H3. Gapdh promoter served as control. NC = non-targeted control, siSetdb1 and siDnmt1 refer to siRNA targeting Setdb1 or Dnmt1, respectively (E). (E) Representative bisulfite sequencing analysis of the Nanog promoter in MEFs cotransduced with OSKM and the indicated combinations on day 15. (F) Model for lincRNA-p21 function in reprogramming. Supportive data are included in Supplementary information, Figure S7.
Collectively, our data imply that lincRNA-p21 preferentially employs a repressive mechanism through H3K9me3 maintenance at the promoters of a subset of pluripotency genes (e.g., Lin28a) and CpG methylation at the promoter of other set of genes (e.g., Nanog), which are both mediated by HNRNPK (Figure 6F).
Discussion
The mammalian genome encodes thousands of lincRNAs, many of which participate in a plethora of biological processes ranging from normal embryonic development to tumorigenesis14. However, only 2 lincRNAs have been reported to promote reprogramming efficiency, lincRNA-RoR and TUNA15,17, whilst lincRNAs that act as a barrier to reprogramming have not yet been identified. Understanding the repertoire of lincRNAs that regulate reprogramming and their roles is relevant because it could help to improve the reprogramming and also shed light on the role of the same lincRNAs in unrelated contexts, particularly in development. Notably, our functional screening has shown that lincRNA-1463 and lincRNA-1526 prevent the preiPSC to iPSC conversion, while lincRNA-1307 facilitates it. These 3 lincRNAs are known for their roles in preventing ESC differentiation along particular lineages22. One possibility is that they funtion by changing the balance of lineage specifiers24,25 that are highly expressed in incompletely reprogrammed colonies12.
We have also shown that a p53-induced lincRNA, lincRNA-p2118, not only sustains the preiPSC state but also poses a barrier for reprogramming. This finding reinforces the notion that p53 impairs reprogramming not exclusively through apoptosis or cell senescence26. Mechanistically, lincRNA-p21 blocks reprogramming by preserving high levels of H3K9me3 and CpG methylation at pluripotency genes. This finding is important, as it may help to understand how lincRNA-p21 represses gene transcription in the DNA damage response18 and potentially other contexts. Moreover, given the role of DNA methylation in sustaining the original cell's epigenetic memory2,27, manipulating lincRNA-p21 expression may help to produce iPSCs of higher quality.
Our results indicate that lincRNA-p21's effects on reprogramming are almost entirely mediated through HNRNPK. In combination, the 2 factors form repressive complexes containing either SETDB1 or DNMT1 at pluripotency gene promoters. However, HNRNPK undoubtedly has additional functions during reprogramming, particularly as it regulates multiple cellular processes including RNA splicing and translational control28. In agreement with this, Hnrnpk knockdown has a more potent enhancing effect on reprogramming compared to lincRNA-p21 knockdown. Likewise, the number of genes regulated by Hnrnpk knockdown in reprogramming exceeds those regulated by lincRNA-p21 knockdown. In this regard, HNRNPK and other members of the hnRNP family interact with multiple lincRNAs29,30. This makes it tempting to speculate that hnRNP proteins are widespread mediators of lincRNA function in reprogramming, which act by facilitating the assembly of various chromatin complexes (activating or repressive) at specific loci. In this scenario, the particular combination of hnRNP proteins and partner lincRNAs at different stages of reprogramming would determine the ultimate effects on this process.
As for the 2 other proteins interacting with lincRNA-p21 identified in our study, it is interesting to note that DNMT1 had been previously shown to interact with non-poly(A) RNAs (lincRNAs are frequently polyadenylated). This mechanism has been proposed to stimulate gene transcription by preventing access of DNMT1 to loci genome-widely31. Our study, in line with another report on Kcnq1ot132, thus expands the concept of DNMT1-interacting RNAs, and suggests that, as opposed to non-poly(A) RNAs, lncRNAs (potentially not just lincRNA-p21 or Kcnq1ot1) promote DNMT1 activity at particular loci.
While this manuscript was in preparation, Dimitrova et al.33 used a genetic deletion approach to demonstrate that lincRNA-p21 also acts in cis to induce p21 expression and diminish MEF proliferation. This contrasts with our work presented here and the work by Huarte et al.18, who could not detect any reduction in p21 expression upon lincRNA-p21 knockdown in MEFs. Dimitrova et al.33 also postulated that lincRNA-p21 blocks OSKM reprogramming of MEFs (measured through alkaline phosphatase staining and qPCR on day 6) by increasing p21, though they did not quantify cell proliferation in this setting. A potential explanation for the discrepancy is that complete suppression (as achieved by genetic deletion) of lincRNA-p21 is necessary to deregulate p21. Additional experimentation will be needed to clarify this issue and it is possible that lincRNA-p21 regulates reprogramming through multiple mechanisms.
Altogether, our study constitutes a resource that may guide others to investigate the functions of lincRNAs in reprogramming, and also provides a new mechanism by which p53, HNRNPK and lincRNA-p21 regulate gene expression. Future work will further delineate the networks controlled by lincRNAs during reprogramming, which will form new connections with the roadblocks identified up to this point.
Materials and Methods
RIP and RNA pull-down
RIP was performed as described34. Briefly, cells were crosslinked with 1% (w/v) formaldehyde (Sigma) and suspended in lysis buffer. Lysates were incubated overnight with the indicated antibodies at 4 °C and RNA-protein complexes were recovered with protein A/G Dynabeads (Life Technologies). RNA pull-down was performed as described34. Briefly, biotinylated lincRNA-p21 and lacZ were synthesized with the mMESSAGE mMACHINE kit (Ambion). Biotinylated RNA was folded and incubated with nuclear extracts. RNA-protein complexes were recovered with Streptavidin agarose beads (Invitrogen). Further details for both procedures are provided in Supplementary information, Data S1.
RNA-seq, bioinformatics and accession number
Sequencing was performed at Guangzhou RiboBio Co., Ltd. with the Illumina HiSeq 2500. RNA-seq data was aligned to the Ensembl v73 transcript annotations using bowtie and RSEM as previously described35. All other bioinformatic analysis was performed using glbase36. These data were deposited in GEO with accession number GSE57967. Further details are provided in Supplementary information, Data S1.
All other methods are described in Supplementary information, Data S1 and Table S3.
Accession codes
References
Takahashi K, Yamanaka S . Induced pluripotent stem cells in medicine and biology. Development 2013; 140:2457–2461.
Liang G, Zhang Y . Genetic and epigenetic variations in iPSCs: potential causes and implications for application. Cell Stem Cell 2013; 13:149–159.
Polo JM, Anderssen E, Walsh RM, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 2012; 151:1617–1632.
Liu L, Xu Y, He M, et al. Transcriptional pause release is a rate-limiting step for somatic cell reprogramming. Cell Stem Cell 2014; 15:574–588.
Rais Y, Zviran A, Geula S, et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature 2013; 502:65–70.
Di Stefano B, Sardina JL, van Oevelen C, et al. C/EBPalpha poises B cells for rapid reprogramming into induced pluripotent stem cells. Nature 2014; 506:235–239.
Guo S, Zi X, Schulz VP, et al. Nonstochastic reprogramming from a privileged somatic cell state. Cell 2014; 156:649–662.
Tapia N, Schöler HR . p53 connects tumorigenesis and reprogramming to pluripotency. J Exp Med 2010; 207:2045–2048.
Soufi A, Donahue G, Zaret KS . Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell 2012; 151:994–1004.
Chen J, Liu H, Liu J, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet 2013; 45:34–42.
Sridharan R, Gonzales-Cope M, Chronis C, et al. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1γ in reprogramming to pluripotency. Nat Cell Biol 2013; 15:872–882.
Mikkelsen TS, Hanna J, Zhang X, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature 2008; 454:49–55.
Subramanyam D, Blelloch R . From microRNAs to targets: pathway discovery in cell fate transitions. Curr Opin Genet Dev 2011; 21:498–503.
Rinn JL, Chang HY . Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012; 81:145–166.
Loewer S, Cabili MN, Guttman M, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet 2010; 42:1113–1117.
Wang Y, Xu Z, Jiang J, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell 2013; 25:69–80.
Lin N, Chang KY, Li Z, et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell 2014; 53:1005–1019.
Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010; 142:409–419.
Yoon JH, Abdelmohsen K, Srikantan S, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell 2012; 47:648–655.
Yang F, Zhang H, Mei Y, Wu M . Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol Cell 2014; 53:88–100.
Silva J, Barrandon O, Nichols J, et al. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol 2008; 6:e253.
Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011; 477:295–300.
Guo X, Liu Q, Wang G, et al. microRNA-29b is a novel mediator of Sox2 function in the regulation of somatic cell reprogramming. Cell Res 2013; 23:142–156.
Montserrat N, Nivet E, Sancho-Martinez I, et al. Reprogramming of human fibroblasts to pluripotency with lineage specifiers. Cell Stem Cell 2013; 13:341–350.
Shu J, Wu C, Wu Y, et al. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell 2013; 153:963–975.
Choi YJ, Lin CP, Ho JJ, et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol 2011; 13:1353–1360.
Lee HJ, Hore TA, Reik W . Reprogramming the methylome: erasing memory and creating diversity. Cell Stem Cell 2014; 14:710–719.
Bomsztyk K, Denisenko O, Ostrowski J . hnRNP K: one protein multiple processes. Bioessays 2004; 26:629–638.
Ng SY, Bogu GK, Soh BS, Stanton LW . The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell 2013; 51:349–359.
Tuck AC, Tollervey D . A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell 2013; 154:996–1009.
Di Ruscio A, Ebralidze AK, Benoukraf T, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature 2013; 503:371–376.
Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C . Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development 2010; 137:2493–2499.
Dimitrova N, Zamudio JR, Jong RM, et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell 2014; 54:777–790.
Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010; 329:689–693.
Pike KA, Hutchins AP, Vinette V, et al. Protein tyrosine phosphatase 1B is a regulator of the interleukin-10-induced transcriptional program in macrophages. Sci Signal 2014; 7:ra43.
Hutchins AP, Jauch R, Dyla M, Miranda-Saavedra D . glbase: a framework for combining, analyzing and displaying heterogeneous genomic and high-throughput sequencing data. Cell Regen 2014; 3:1.eCollection 2014.
Acknowledgements
We thank all members of the Esteban Lab for their support. This work was supported by the National Basic Research Program of China (973 Program; 2011CB965201 and 2011CBA01106), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA01020106), the National Natural Science Foundation of China (31371513 and 81202010), the Cooperation Program of the Research Grants Council (RGC) of the Hong Kong Special Administrative Region and the National Natural Science Foundation of China (81261160506), the RGC of the Hong Kong Special Administrative Region, China (476310), the Natural Science Foundation of Guangdong Province (S2013010015487), the Bureau of Science, Technology and Information of Guangzhou Municipality (2012J5100040) and the International Science and Technology Cooperation Program of China (2013DFE33080).
Author information
Authors and Affiliations
Corresponding authors
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Table S1
LincRNA expression in 4 cell types and reprogramming (related to Figure 1 and 2, Supplementary information, Figure S1 and S2) (XLSX 29 kb)
Supplementary information, Table S2
RNA-seq gene list (fold change higher than 1.5) at the indicated time points with lincRNA-p21 or Hnrnpk knockdown (related to Figure 5, Supplementary information, Figure S5 and S6). (XLSX 569 kb)
Supplementary information, Table S3
shRNA sequences, primers and antibodies used in this study (XLSX 29 kb)
Supplementary information, Figure S1
Expression of lincRNAs in reprogramming (related to Figure 1) (PDF 153 kb)
Supplementary information, Figure S2
LincRNA-p21 is induced by p53 (related to Figure 2) (PDF 321 kb)
Supplementary information, Figure S3
Comparison of cell proliferation and reprogramming efficiency in reprogrammed MEFs with lincRNA-p21 or/and p53 knockdown and iPSC clone characterization (related to Figure 2) (PDF 455 kb)
Supplementary information, Figure S5
LincRNA-p21 is a subset of HNRNPK function in reprogramming (related to Figure 5) (PDF 1289 kb)
Supplementary information, Figure S6
LincRNA-p21 influences the maturation of let-7 miRNAs but not the expression of mesenchymal or epithelial genes (related to Figure 5) (PDF 391 kb)
Supplementary information, Figure S7
HNRNPK binds to pluripotency genes in reprogramming and lincRNA-p21 regulates CpG methylation at the Nanog promoter (related to Figure 6) (PDF 479 kb)
Supplementary information, Data S1
Extended Materials and Methods (PDF 261 kb)
Rights and permissions
About this article
Cite this article
Bao, X., Wu, H., Zhu, X. et al. The p53-induced lincRNA-p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promoters. Cell Res 25, 80–92 (2015). https://doi.org/10.1038/cr.2014.165
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2014.165
Keywords
This article is cited by
-
Non-coding RNA regulation of integrins and their potential as therapeutic targets in cancer
Cellular Oncology (2023)
-
LncRNA LYPLAL1-AS1 rejuvenates human adipose-derived mesenchymal stem cell senescence via transcriptional MIRLET7B inactivation
Cell & Bioscience (2022)
-
5mC and H3K9me3 of TRAF3IP2 promoter region accelerates the progression of translocation renal cell carcinoma
Biomarker Research (2022)
-
CRMP2 derived from cancer associated fibroblasts facilitates progression of ovarian cancer via HIF-1α-glycolysis signaling pathway
Cell Death & Disease (2022)
-
p63 silencing induces epigenetic modulation to enhance human cardiac fibroblast to cardiomyocyte-like differentiation
Scientific Reports (2022)