Abstract
Trans-splicing, a process involving the cleavage and joining of two separate transcripts, can expand the transcriptome and proteome in eukaryotes. Chimeric RNAs generated by trans-splicing are increasingly described in literatures. The widespread presence of antibiotic resistance genes in natural environments and human intestines is becoming an important challenge for public health. Certain antibiotic resistance genes, such as ampicillin resistance gene (Ampr), are frequently used in recombinant plasmids. Until now, trans-splicing involving recombinant plasmid-derived exogenous transcripts and endogenous cellular RNAs has not been reported. Acyl-CoA:cholesterol acyltransferase 1 (ACAT1) is a key enzyme involved in cellular cholesterol homeostasis. The 4.3-kb human ACAT1 chimeric mRNA can produce 50-kDa and 56-kDa isoforms with different enzymatic activities. Here, we show that human ACAT1 56-kDa isoform is produced from an mRNA species generated through the trans-splicing of an exogenous transcript encoded by the antisense strand of Ampr (asAmp) present in common Ampr-plasmids and the 4.3-kb endogenous ACAT1 chimeric mRNA, which is presumably processed through a prior event of interchromosomal trans-splicing. Strikingly, DNA fragments containing the asAmp with an upstream recombined cryptic promoter and the corresponding exogenous asAmp transcripts have been detected in human cells. Our findings shed lights on the mechanism of human ACAT1 56-kDa isoform production, reveal an exogenous-endogenous trans-splicing system, in which recombinant plasmid-derived exogenous transcripts are linked with endogenous cellular RNAs in human cells, and suggest that exogenous DNA might affect human gene expression at both DNA and RNA levels.
Similar content being viewed by others
Introduction
Trans-splicing involves the cleavage and joining of two separate transcripts, and is a novel mode of alternative splicing by which multiple mRNAs and proteins with different functions are produced from a single gene; this can expand the transcriptome and proteome in eukaryotes1,2,3. Trans-splicing was first discovered as the spliced leader-addition trans-splicing in trypanosomes4,5 and later reported in higher eukaryotes, such as flies6,7, rats8,9 and humans10,11,12,13,14. Trans-splicing is increasingly reported by experimental observations or through computational analyses15,16,17,18,19. Based on identified cases in mammalian species, trans-splicing can be classified into several categories: intragenic, intergenic and interchromosomal trans-splicing2. The duplications of exons in trans-spliced mRNAs of the rat carnitine octanoyltransferase gene (COT) and the human estrogen receptor-α gene (hERα) are examples of intragenic trans-splicing8,9,20. In human liver, the production of a variety of hybrid mRNAs derived from a cluster of P450 3A genes is an example of intergenic trans-splicing12. The chimeric RNA that contains exons of the JAZF1 gene located on chromosome 7 and the JJAZ1 gene located on chromosome 17 is an example of interchromosomal trans-splicing14. Trans-splicing of endogenous cellular RNAs and exogenous transcripts, however, has not yet been reported.
Antibiotic resistance genes are increasingly prevalent in both benign and pathogenic bacteria, posing an emerging threat to public health21. The continued evolution and widespread dissemination of antibiotic resistance genes, and their acquisition by bacterial pathogens are becoming one of the most important challenges for clinical medicine22. For example, the exchange of antibiotic resistance genes between environmental bacteria and clinical pathogens was recently reported23. Natural environments and human intestines represent important reservoirs of antibiotic resistance genes24,25. In particular, certain antibiotic resistance genes such as ampicillin resistance gene (Ampr) are frequently used in recombinant plasmids. Recombinant plasmids were created in the early 1970s26,27 and have been extensively used in research, agriculture and medicine for nearly four decades. The most common application of recombinant plasmids is in basic molecular biology and molecular medicine research28. In addition, recombinant plasmids have been used in transgenic technology for agricultural purposes29. Recombinant proteins, the expression products of recombinant genes, are also widely used in pharmacological and medical fields30. However, studies on the presence of antibiotic resistance genes or recombinant plasmid-derived fragments in human tissues or cells are rare.
The family of acyl-coenzyme A:cholesterol acyltransferase (ACAT), also known as sterol O-acyltransferase, consists of two different enzymes (ACAT1 and ACAT2) and catalyzes the formation of cholesteryl esters from cholesterol and long-chain fatty acyl-coenzyme A. Both ACATs are membrane-bound acyltransferases, employing histidine and asparagine as the two active-site residues31, and play important roles in cellular cholesterol homeostasis32. ACAT1 is a homotetrameric protein located at the endoplasmic reticulum with 9 transmembrane domains33,34,35. Human ACAT1 cDNA K1 was identified by its ability to complement the functional deficiency of mutant CHO cells lacking ACAT activity36. K1 cDNA is 4 011 bp in length, contains an open reading frame (ORF) of 1 650 bp encoding a protein of 550 amino acids, and has a long 5′-UTR of 1 396 bp. ACAT1 genomic DNA contains 18 exons (exons Xa, Xb and 1-16); exons 1-16 are located on chromosome 1, whereas exon Xa (1 279 bp) is located on chromosome 710. The origin of the mini-exon Xb of 10 bp is still unclear at present.
Northern blot analysis has revealed the presence of four ACAT1 transcripts (7.0, 4.3, 3.6 and 2.8 kb) in all of the human tissues and cell lines examined, with the 3.6- and 2.8-kb mRNAs being the most prevalent. The 4.3-, 3.6- and 2.8-kb mRNAs share the sequence derived from chromosome 1, but only the 4.3-kb mRNA contains the long exon Xa located on chromosome 7. This suggests that the 4.3-kb transcript is derived from two different chromosomes, presumably by an interchromosomal trans-splicing event10. The 3.6- and 2.8-kb mRNAs only produce the ACAT1 50-kDa isoform, whereas the 4.3-kb chimeric mRNA produces two ACAT1 isoforms (50-kDa and 56-kDa) using AUG1397-1399 and its upstream in-frame GGC1274-1276 as the presumed initiation codon, respectively37,38. The 56-kDa isoform is expressed in human THP-1 macrophages and human monocyte-derived macrophages37.
Ectopic expression studies have shown that the 56-kDa isoform is enzymatically active, but its normalized activity is only approximately 30% of the activity of the 50-kDa isoform37. When the 50- and 56-kDa isoforms were coexpressed ectopically in cultured cells, the normalized ACAT1 activity was approximately 50% of the activity of the 50-kDa isoform expressed alone37. The presence of an RNA secondary structure (nt 1 355-1 384) within the long 5′-UTR enhances the selection of the downstream AUG1397-1399 as the initiation codon to produce the 50-kDa isoform39, whereas the two upstream RNA secondary structures (nt 1 255-1 268 and nt 1 286-1 342), involving sequences transcribed from two different chromosomes, are required to produce the 56-kDa isoform38. To our knowledge, no analogues of human ACAT1 chimeric mRNA have been found in other animals, including nonhuman primates and rodents.
In the current work, through mass spectrometry (MS) analysis and protein N-terminal amino acid sequencing, we found that the human ACAT1 56-kDa isoform contains an extra N-terminal region of 46 amino acids that are identical to that encoded by the antisense strand of Ampr (asAmp) present in common recombinant Ampr-plasmids. Moreover, DNA fragments containing the asAmp with an upstream cryptic promoter and the corresponding exogenous asAmp transcripts have been detected in human blood cells and cell lines. We have also demonstrated that the exogenous asAmp transcript with a typical AUG start codon is trans-spliced to the endogenous 4.3-kb human ACAT1 chimeric mRNA in a canonical manner, indicating that the human ACAT1 56-kDa isoform is produced via an exogenous-endogenous (exo-endo) trans-splicing event.
Results
Human ACAT1 56-kDa isoform contains an extra N-terminal region encoded by asAmp derived from common recombinant Ampr-plasmids
We previously demonstrated that the 4.3-kb human ACAT1 chimeric mRNA (corresponding to cDNA K1) produces 50- and 56-kDa isoforms by using the typical start codon AUG1397-1399 and its upstream in-frame codon GGC1274-1276 as a presumed start codon, respectively37,38. To verify the presumed role of the GGC1274-1276 codon in translation initiation, we generated various mutations within this codon in the full-length (4 011 bp) ACAT1 cDNA K1 (Figure 1A, top panel) and performed western blot analyses using an anti-ACAT1 antibody that recognizes both 50- and 56-kDa isoforms (Figure 1A, bottom panel) after ectopic expression of the constructs in AC29 cells, an ACAT1-deficient mutant CHO cell line.
An extra N-terminal region contained in human ACAT1 56-kDa isoform is encoded by a sequence present in recombinant Ampr-plasmids. (A-C) Western blot analysis of the expression of WT and mutant ACAT1 constructs. Constructs (numbers 1-23) containing the entire ACAT1 (A), partial ACAT1 (B, C) or Rluc (C) were transfected separately into AC29 cells. Western blot analyses were performed with anti-ACAT1 or anti-Rluc antibodies. Filled circle, GGC1274-1276 codon; hollow circle, ATG1397-1399 codon; and underlined characters, mutated nucleotides. (D) Calculated molecular weights (MW) of proteins initiated from the GGC1274-1276, ATG1274-1276 and ATG1397-1399 codons. ACAT1-50NT, 130 amino acids encoded by the sequence from ATG1397-1399 to GAT1784-1786. ACAT1-56uNT, 41 amino acids upstream of ACAT1-50NT. Filled circle, glycine; hollow circle, methionine; and aa, amino acids. (E) Purification and 2-D electrophoresis of the 26-kDa ACAT1 protein. Construct pNTF-number 10 (B) was expressed in AC29 cells to generate the 26-kDa ACAT1 protein. The proteins were purified with ANTI-FLAG M2 Affinity Gel and separated by 2-D electrophoresis. Arrows indicate spots of different 26-kDa proteins designated as P1, P2 and P3. PCMV, CMV promoter (black arrow); ori, ColE1 origin (gray arrow). (F) MALDI-TOF MS analysis of the purified 26-kDa proteins. Mass spectra are recorded in the positive mode and listed for the purified 26-kDa protein P2. (G) The N-terminal region of the purified P2 protein and its origin. The 15 amino acids (bold) were determined by the N-terminal sequencing of the purified P2 protein; they are encoded by the underlined sequence that matches the sequence of asAmp derived from recombinant plasmids (asAmp, tessellated box). The amino acid sequence of asAmp-encoding p88 protein (asAmp-P88) and the region of a recombined cryptic promoter are shown. (H) Schematic representation of human ACAT1-56NT region. Italics indicate the N-terminal 15 amino acids of the purified P2 protein. Asterisks indicate modifications by iodoacetamide on cysteines. Trypsin cleavage sites are indicated by bold characters and calculated MWs of representative peptides (underlined) after trypsin cleavage are shown. ACAT1-56eNT, 43 or 46 amino acids from the asAmp-P88. (I) Activity of a recombined cryptic promoter upstream of the asAmp ORF region. Reporter plasmids containing the luciferase gene under the control of various-sized upstream sequences of the asAmp ORF region were transfected into HEK293 cells, and their relative luciferase activities were determined. Means and SDs are shown (n = 3). Value with the promoter-less plasmid is defined as 1.0.
Unexpectedly, the western blot results showed that when the GGC1274-1276 codon was mutated to the typical initiation codon ATG, an additional medium-sized protein band (approximately 53 kDa) appeared in addition to the larger (56 kDa) and smaller (50 kDa) isoforms (number 9, Figure 1A, bottom panel). To further verify the result, we constructed two series of expression plasmids containing ACAT11243-1786 linked to 3× Flag sequence (Figure 1B, top panel) or ACAT11243-1396 linked to the Renilla luciferase (Rluc) ORF (Figure 1C, top panel). These constructs also each contain an artificially introduced in-frame ATG codon at, or upstream or downstream of the GGC1274-1276 position (Figures 1B and 1C, top panel). Western blot analysis showed that all constructs with the introduced ATG codon produced additional medium-sized proteins (approximately 22 kDa in Figure 1B, or 40 kDa in Figure 1C) in addition to the corresponding larger (26 kDa in Figure 1B or 44 kDa in Figure 1C) and smaller (18 kDa in Figure 1B, or 36 kDa in Figure 1C) proteins. Interestingly, the larger hybrid protein disappeared when CAG1271-1273 was replaced with the ATG codon (Figure 1B, number 13 and Figure 1C, number 20).
Based on the results shown in Figure 1B and 1C, we calculated the protein molecular weights. As shown in Figure 1D, calculated molecular weights of the proteins initiated from the presumed start codon GGC1274-1276 (22.59 kDa of Gly-ACAT1-56uNT-50NT-Flag and 40.09 kDa of Gly-ACAT1-56uNT-Rluc) were less than the sizes of the larger protein bands observed in western blot analyses (26 kDa in Figure 1B and 44 kDa in Figure 1C), whereas the calculated molecular weights of the medium-sized proteins initiated from the introduced ATG codon (22.67 kDa of Met-ACAT1-56uNT-50NT-Flag and 40.17 kDa of Met-ACAT1-56uNT-Rluc) and the smaller proteins initiated from the typical start codon ATG1397-1399 (18.45 kDa of Met-ACAT1-50NT-Flag and 36.00 kDa of Met-Rluc) were fully consistent with the sizes observed in Figure 1B and 1C. These results suggest that the human ACAT1 56-kDa isoform may not be initiated from the GGC1274-1276 codon, and most likely it contains an extra N-terminal region in front of the 41 amino acids (designated as ACAT1-56uNT in Figure 1D) encoded by the sequence from GGC1274-1276 to AUG1397-1399.
To identify the sequence of the possible extra N-terminal region of the human ACAT1 56-kDa isoform, the expression plasmid pNTF (Figure 1B, number 10, and Figure 1E) was used and the expressed proteins were precipitated using anti-Flag antibodies, seperated by 2-D electrophoresis, and stained with Coomassie blue dye. The 26-kDa proteins produced three apparent spots, designated as P1, P2 and P3 (Figure 1E). The purified protein samples from these three spots were analyzed individually by MALDI-TOF MS and their N-terminal amino acids were identified by peptide sequencing. MS results showed that three specific tryptic fragments with sizes of 1493.626, 1821.932 and 2241.867 were present in all three spots (Figure 1F and Supplementary information, Figure S1). The size of the largest tryptic fragment (2241.867) as determined by the MS analysis was consistent with the calculated molecular weight (2242.07) of a predicted tryptic fragment within human ACAT1-56uNT (Figure 1H, underlined), confirming the observation in our previous work37.
However, only the sample from spot P2, but not P1 or P3 (which possibly contains the closed N-terminus due to unidentified modification), could be sequenced; the sequencing results reveal that its N-terminal 15 amino acids were MIPRDPRSPAPDLSA. Surprisingly, this sequence matched 100% with the N-terminal amino acids of asAmp-P88 (Figure 1G, bold) encoded by the 267-bp ORF of asAmp that is present in recombinant Ampr-plasmids (Figure 1G). In addition, the calculated molecular weights (1493.64 and 1821.95) of two predicted tryptic fragments of the asAmp-P88 protein (Figure 1H, underlined) were consistent with the sizes (1493.626 and 1821.932) of the two observed smaller tryptic fragments in MS analysis (Figure 1F).
These data demonstrate that the human ACAT1 56-kDa isoform contains an extra N-terminal region (designated as ACAT1-56eNT in Figure 1H) that corresponds to the N-terminal region of asAmp-P88, and also suggest that the Gly residue encoded by the GGC1274-1276 codon which is not the start codon as previously presumed, is linked to an Arg residue of asAmp-P88. Based on the molecular weight difference (about 4 kDa, Figure 1B) between the larger- and medium-sized protein bands, we reasoned that the ACAT1-56eNT should be of 43 or 46 amino acids (Figure 1H, dashed line). Moreover, a recombined cryptic promoter (RP) with apparent biological activity in higher eukaryotic cells (Figure 1I) was identified upstream of the asAmp DNA in a region common to many recombinant Ampr-plasmids (Figure 1G) such as pBR322 and pcDNA3. Therefore, it is likely that the exogenous asAmp can be transcribed in cells transfected with pcDNA3-derived recombinant Ampr-plasmids.
Considering that the ACAT1-56NT is likely composed of peptides derived from asAmp-P88 and ACAT1, we hypothesized that the human ACAT1 56-kDa isoform might be the product of a trans-spliced mRNA generated from the endogenous cellular ACAT1 mRNA and the exogenous asAmp transcript derived from common recombinant Ampr-plasmids. Hence, it would be important to determine whether the exogenous asAmp DNA fragments with an upstream cryptic promoter derived from recombinant plasmids and the corresponding transcripts exist in human cells.
The exogenous asAmp DNA and the corresponding transcripts exist in human blood cells
We previously reported that the ACAT1 56-kDa isoform is expressed in human THP-1 macrophages and human monocyte-derived macrophages37. To examine the potential presence of exogenous asAmp DNA fragments in human cells, samples of peripheral blood mononuclear cells (PBMC) from 50 healthy people were randomly collected for detailed characterization. The samples were first examined to rule out contamination of low-molecular-weight plasmids or bacteria (Supplementary information, Figure S2). The relative quantity of asAmp DNA in adherent or non-adherent cells of each sample was determined by real-time quantitative PCR (qPCR) (Figure 2A).
Expression of the asAmp transcripts and ACAT1 56-kDa isoform in human blood cells. (A-D) qPCR analysis of genomic DNA from 50 human PBMC samples. Adherent and non-adherent cells were separated from the PBMC samples. The quantities of asAmp and ACAT1 DNA in adherent and non-adherent cells were determined by qPCR with specific primer sets asAmp-F/asAmp-R and A1C7-F/A1C7-R, complementary to the sequence of asAmp and human chromosome 7-derived ACAT1 (A1C7), respectively. The relative copy number of asAmp DNA per cell was determined by normalizing the DNA quantity of asAmp to that of ACAT1(set as single copy per cell). The relative asAmp DNA copy number per cell of individual samples is shown in A. (B) The relative asAmp DNA copy numbers of the PBMC samples were compared between the adherent and non-adherent cells, and the statistical significance was evaluated using a two-tailed Student's t-test (n = 50; ***P < 0.001). (C, D) Pie charts illustrate the percentage of the samples with relative asAmp DNA copy ≥ 1 or < 1 in adherent (C) and non-adherent (D) cells. (E-I) Analysis of four paired adherent (a) and non-adherent (na) cells from four different PBMC samples. (E) Genomic DNAs were analyzed by PCR with specific primer sets including asAmp-F/asAmp-R, A1C7-F/A1C7-R (as those used in A), RP-F/asAmp-R (complementary to the DNA sequence of asAmp with an upstream recombined cryptic promoter from Ampr-plasmids), and Kanr-F/Kanr-R (complementary to the sequence of kanamycin resistance gene). (F) The transcribed RNA and putative trans-spliced products were analyzed by RT-PCR with specific primer sets including asAmp-F/asAmp-R (as that used in A) and asAmp-F/A1-R1 (complementary to the sequences of asAmp and ACAT1, respectively). GAPDH served as the internal control. (G, H) The RT-PCR products shown in F were further subjected to EcoRI digestion (G) and DNA sequencing (H). The determined DNA sequence contains sequences of both asAmp and ACAT1. Several amino acid codons, the trans-splicing junction (dashed line) and partial sequences of exon 1, exon Xa and mini-exon Xb (underlined) containing EcoRI site (bold) are indicated in H. (I) Western blot was performed to detect the ACAT1 isoforms with anti-asAmp-P88 or anti-ACAT1 antibodies. Actin served as an internal control. NC, negative-control without any templates or using lysates of unmanipulated AC29 cells. PC, positive-control using lysates of AC29 cells transfected with pA.
The results show that the relative copy number of asAmp DNA per cell was significantly higher in adherent cells than in non-adherent cells (Figure 2B). A higher abundance of asAmp DNA (≥ 1 copy per cell) was detected in the adherent cells in approximately 30% of the samples (Figure 2C), 15-fold higher than in non-adherent cells (2%) (Figure 2D). The variation of the asAmp DNA quantity among individual blood samples and between adherent and non-adherent cell types may relate to the health status of individuals in their respective environments and the abilities of their phagocytes including macrophages (which would mostly appear as adherent cells) to obtain the asAmp DNA or recombinant Ampr-plasmids from phagocytized foreign substances.
Furthermore, four representative PBMC samples (Figure 2A, oval framed) were analyzed by PCR, RT-PCR and western blot analyses. The PCR assays analyzing the genomic DNA (Figure 2E) revealed that DNA fragments of asAmp (top panel) or asAmp with an upstream recombined cryptic promoter derived from Ampr-plasmids (RP-asAmp, secondary panel) were present in cells with higher quantity of asAmp DNA (approximately one copy per cell as shown in Figure 2A). DNA sequencing further verified that the PCR products of the RP-asAmp fragments contain sequences from both the asAmp and its upstream recombined cryptic promoter (Supplementary information, Figure S3). The partial ACAT1 DNA fragment derived from chromosome 7 was amplified in parallel as a control (Figure 2E, third panel). No amplicons of the kanamycin resistance gene (Kanr) were observed (Figure 2E, bottom panel). These results suggest that human cells harbor certain DNA fragments derived from Ampr-plasmids in their genomes, likely as a result of the spreading of the plasmids through their bacterial hosts. Nevertheless, the precise routes of the spreading remain to be determined.
Importantly, the asAmp transcripts and putative trans-spliced products of the asAmp transcripts and ACAT1 mRNAs were identified by RT-PCR (Figure 2F), and the characteristic digestion with EcoRI (Figure 2G). Moreover, DNA sequencing of the RT-PCR products indicated that the plasmid-derived exogenous asAmp transcript was trans-spliced with the 4.3-kb endogenous human ACAT1 chimeric mRNA, and that the AGA codon (coding for R46) of the asAmp transcript was linked to the GGC1274-1276 codon (coding for G) of human ACAT1 mRNA (Figure 2H). As the 4.3-kb human ACAT1 chimeric mRNA is derived from two different chromosomes (7 and 1) presumably by an interchromosomal trans-splicing event, it is impossible that the final spliced product with the determined sequence (Figure 2H) results from the processing of a pre-mRNA containing both the integrated asAmp sequence and the 4.3-kb ACAT1 chimeric mRNA sequence by cis-splicing. Accordingly, western blot analyses using anti-asAmp-P88 (which recognizes amino acids 19-36 of asAmp-P88) or anti-ACAT1 (which recognizes the N-terminal 125 amino acids of the ACAT1 50-kDa isoform) antibodies demonstrated that the human ACAT1 56-kDa isoform was present in the PBMC samples (Figure 2I), which is consistent with the observations from our previous work37.
Collectively, these results suggest that the ACAT1 56-kDa isoform is produced in human blood cells from a trans-spliced mRNA containing the plasmid-derived exogenous asAmp sequence and the 4.3-kb human ACAT1 chimeric mRNA sequence, and that exogenous DNA fragments could be incorporated into the human genome and affect human gene expression at both DNA and RNA levels.
The exogenous asAmp transcripts are broadly present in higher eukaryotic cells
The results described above reveal that asAmp transcripts derived from recombinant Ampr-plasmids can be expressed in the blood cells of healthy individuals. We next sought to check the expression of asAmp transcripts in cell lines that are used extensively in biological research. We performed PCR assays to analyze the genomic DNAs of nine human cell lines, and found that asAmp and RP-asAmp DNAs were amplified in these cell lines (Figure 3A, top and secondary panels). A DNA fragment of the Kanr gene was also detected in HeLa cells (Figure 3A, fourth panel). To rule out the possibility of microbe contaminations, all genomic DNA samples were examined by PCR using primers for a universally conserved bacterial gyrB gene40; no bacterial DNA was detected (Figure 3A, bottom panel), further confirming that DNA fragments of asAmp and RP-asAmp exist in these cell lines.
Existence of asAmp transcripts in higher eukaryotic cells. (A) PCR analysis of genomic DNAs from various human cell lines. Primer sets asAmp-F/asAmp-R, RP-F/asAmp-R, A1C7-F/A1C7-R and Kanr-F/Kanr-R are the same as those in Figure 2E. Primer set gyrB-F/gyrB-R is complementary to the sequence of bacterial gyrB cDNA. NC, negative-control without DNA templates. (B) RT-PCR analysis of asAmp transcripts in human cell lines HEK293T and THP-1. Primer set asAmp-F/asAmp-R was used. NC, without RNA templates. (C) The PCR products from HEK293T cells (B) were subjected to DNA sequencing. The sequence matching the 267-bp asAmp ORF is shown. (D, E) Alignment analysis of the asAmp ORF in EST database. The 267-bp asAmp ORF sequence is entered as query sequence by using BLASTN. (D) Results are shown by taxonomy tree with hit numbers in each species indicated in brackets. (E) The species containing the 267-bp asAmp ORF (gray bar) with 100% identity. Branch lengths are not proportional to phylogeny time.
Consistently, asAmp transcripts were detected in HEK293T and THP-1 cells by RT-PCR (Figure 3B), and the results of DNA sequencing showed that the sequence of the asAmp transcript in HEK293T cells was identical to that of asAmp present in recombinant plasmids (Figure 3C). Moreover, the asAmp ORF sequence (267 bp) was found in the EST database (dbEST) using the BLASTN (http://blast.ncbi.nlm.nih.gov/)41. The results of the BLASTN analysis showed that the asAmp cDNAs have been cloned from various higher eukaryotic cells, including both plant and animal cells (Figure 3D). In particular, cDNA sequences corresponding to the asAmp-P88 ORF (267 bp) found in a number of species matched perfectly with each other (with 100% sequence identity) (Figure 3E). These data demonstrate that the exogenous asAmp transcripts derived from Ampr-plasmids are broadly present in higher eukaryotic cells.
The asAmp transcripts can be trans-spliced to human ACAT1 mRNA in intact-cell and cell-free systems
As shown in Figure 2, the trans-splicing of asAmp and ACAT1 transcripts occurs in the blood cells of healthy individuals. Next, experiments to further probe this trans-splicing process were carried out in intact-cell and cell-free systems. First, several constructs were generated based on a Kanr-plasmid vector pVk (Ampr-free, Figure 4A, number 24); these include plasmids containing the asAmp-P88 ORF sequence and plamids containing ACAT11-1786, all of which contain Flag or Myc sequences at either 5′-end of asAmp or 3′-end of ACAT1 (Figure 4A, numbers 25-28). These expression plasmids, together with the Ampr-plasmid pcDNA3 (Figure 4A, number 29), were transfected separately or in different combinations into AC29 cells. RT-PCR and western blot analyses were then performed.
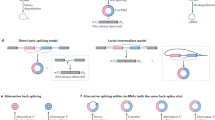
Exo-endo trans-splicing of the asAmp and human ACAT1 transcripts. (A) Schematic representation of constructed plasmids (numbers 25-28) containing asAmp or ACAT11-1786 cDNA fused to Flag or Myc tags and empty vectors (numbers 24 and 29) with (pcDNA3) or without (pVk) the Ampr. (B, C) Analysis of trans-spliced RNA products and their translated proteins in intact cells. (B) AC29 cells were transfected with the indicated plasmids. RT-PCR was performed with primer sets asAmp-F/asAmp-R, asAmp-F/A1-R1, A1-F1/A1-R1 to detect the expression of asAmp, trans-spliced asAmp-ACAT1 and ACAT1, respectively. (C) Anti-Flag or anti-Myc antibodies were used in western blot analyses. Calculated MWs of various fusion proteins are shown. Arrows indicate fusion proteins identified by both anti-Flag and anti-Myc antibodies. (D) Schematic representation of in vitro-prepared transcripts as 5′-donor (D1) and 3′-acceptors (A1 and A2). (E) Analysis of trans-spliced RNA in the cell-free system. The cell-free trans-splicing reaction was performed using HeLa cell nuclear extract (NE) and in vitro-prepared 5′-donor and 3′-acceptors depicted in D. Specific primer set asAmp-F/A1-R2 was used in the RT-PCR assay. asAmp-F is the same as in C; A1-R2 is complementary to the sequence of ACAT1 cDNA (upstream of A1-R1); NC, negative-control without templates. (F) DNA sequencing of the RT-PCR product (lane 2) from E. (G) Splicing efficiency of the in vitro trans-splicing between asAmp and ACAT1 RNAs was determined by qRT-PCR assays. The amount of spliced and unspliced RNA products were determined with specific primer sets asAmp-F/A1-R2 (as that used in E) and A1-F2/A1-R2 (complementary to the sequence of ACAT1 cDNA). The percentage of spliced RNAs was calculated by dividing the amount of spliced RNAs by the total RNAs. Means and SD are shown (n = 3). (H) Schematic representation of the exo-endo trans-splicing between asAmp and ACAT1 RNAs and the production of ACAT1 56-kDa/50-kDa isoforms. Human ACAT1 56-kDa isoform is the product of the exo-endo trans-splicing of exogenous asAmp and endogenous ACAT1 transcripts, and the ACAT1 50-kDa isoform is translated by using AUG1397-1399 as the start codon. Underlined characters, AUG start codon; bold characters, AGA and GGC codons at the junction site; italic characters, splice-site signals (5′ SS and 3′ SS).
The results of RT-PCR showed that the asAmp transcripts (Figure 4B, top panel, lanes 1-3 and 5-7) and the human ACAT1 mRNA (Figure 4B, bottom panel, lanes 1-8) were present in cells transfected with the corresponding plasmids. In addition, trans-spliced mRNAs were detected by RT-PCR in cells expressing both asAmp and human ACAT1 transcripts (Figure 4B, middle panel, lanes 1-3 and 5-7). Importantly, results of western blot analyses using anti-Flag and anti-Myc antibodies demonstrated that the sizes of the hybrid proteins were in accordance with the calculated molecular weights (37.0, 35.9, 32.7, 31.8, 30.7 and 27.5 kDa), and that these hybrid proteins were detected in cells with ectopic expression of both asAmp and human ACAT1 transcripts (Figure 4C, lanes 1-3 and 5-7), but not in cells merely transfected with the plasmids expressing the fused protein of ACAT1-50NT-3Flag (18.5 kDa) or ACAT1-50NT-6Myc (23.7 kDa) (Figure 4C, lanes 4 and 8). In particular, hybrid proteins 3Myc-ACAT1-56eNT-56uNT-50NT-3Flag (31.8 kDa) and 3Flag-ACAT1-56eNT-56uNT-50NT-6Myc (35.9 kDa) were simultaneously identified by both anti-Flag and anti-Myc antibodies (Figure 4C, lanes 2 and 5, arrows), confirming that these proteins were translated from trans-spliced mRNAs.
All of the results described above were obtained in intact cells. We next used a cell-free splicing assay to test the trans-splicing of asAmp and ACAT1 RNAs. Three RNA substrates, including a 5′-donor of a full-length asAmp transcript (D1) and two 3′-acceptors corresponding to different regions of human ACAT1 mRNA (A1 and A2), were prepared by in vitro transcription (Figure 4D). All RNA substrates were examined by denaturing polyacrylamide gel electrophoresis to ensure their identities as pre-RNAs (Supplementary information, Figure S4). The RNA substrates were incubated with nuclear extract (NE) from HeLa cells under optimal splicing conditions42,43 and the spliced products were examined by RT-PCR. As demonstrated in Figure 4E, the trans-spliced RNA products were detected when D1 was incubated in NE with either A1 or A2 (Figure 4E, lanes 2 and 3), whereas no product was observed when NE alone was present (Figure 4E, lane 1). Moreover, DNA sequencing of the RT-PCR products indicated that the asAmp RNA was trans-spliced to human ACAT1 RNA substrate in vitro (Figure 4F), which is consistent with the results obtained in intact human blood cells (Figure 2H). The efficiency of the in vitro splicing reaction was determined by quantitative RT-PCR assays. As shown in Figure 4G, about 1.5% of the added RNA substrates were spliced 90 min after the beginning of the reaction, which is comparable to the efficiency reported in literatures42,43.
These results demonstrate that the trans-splicing between the asAmp and the human ACAT1 transcripts occurs in a cell-free system. According to the identified sequence of the trans-spliced RNA product (Figure 4H, top panel), the AGA136-138 codon (bold) of the asAmp transcript connects with the GGC1274-1276 codon (bold) of human ACAT1 mRNA, and the sequences adjacent to these two codons (GUAAGU and CAG, italic) are classical splice-site signals (5′ SS and 3′ SS). As one RNA substrate is an exogenous transcript derived from recombinant plasmids and the other represents an endogenous cellular mRNA, we name this novel trans-splicing reaction exogenous-endogenous (exo-endo) trans-splicing in human cells (Figure 4H). In addition, our data indicate that the translation of the human ACAT1 56-kDa isoform begins from the typical AUG codon of the asAmp transcript, whereas the translation of the 50-kDa isoform is initiated from the AUG1397-1399 codon contained in the 4.3-kb human ACAT1 chimeric mRNA (Figure 4H).
The exo-endo trans-splicing requires the specific splice-site signals
As shown in Figure 1B and 1C, larger proteins were no longer produced and medium-sized proteins appeared when CAG1271-1273 was replaced with the ATG codon (pNTF-M18, number 13 in Figure 1B and pNTR-M18, number 20 in Figure 1C), which supports our hypothesis that CAG1271-1273 acts as a splice-site signal for the trans-splicing of asAmp and ACAT1 transcripts. To further verify this, the Ampr-plasmids pNTF, pNTF-M18 (Figure 5A), pNTR and pNTR-M18 (Figure 5B) were transfetced into AC29 cells followed by western blot and RT-PCR analyses. Western blot analyses using anti-ACAT1 and anti-Rluc antibodies (Figures 5A and 5B, left of middle panel) confirmed that medium-sized proteins (22 and 40 kDa), but not larger proteins (26 and 44 kDa) were produced when CAG1271-1273 was mutated to ATG. Moreover, western blot analyses using an anti-asAmp-P88 antibody (Figures 5A and 5B, right of middle panel) and RT-PCR assays using specific primers (Figures 5A and 5B, bottom panel) detected the existence of larger proteins harboring ACAT1-56eNT (N-terminal 46 amino acids of asAmp-P88) and their corresponding trans-spliced mRNAs, respectively, in cells transfected with pNTF (number 10) or pNTR (number 17) containing wild-type (WT) CAG1271-1273, but not in cells transfected with pNTF-M18 (number 13) or pNTR-M18 (number 20) containing the ATG codon as a substitution for CAG1271-1273. These results further support that CAG1271-1273 of the human ACAT1 transcript specifically functions as a 3′ SS.
The 5′ SS and 3′ SS are required for the exo-endo trans-splicing. (A, B) Analysis of the products from the exo-endo trans-splicing event by western blot and RT-PCR assays. Anti-asAmp-P88, anti-ACAT1 and anti-Rluc antibodies were employed in the western blot analyses. Specific primer sets asAmp-F/A1-R1 (A) and asAmp-F/RL-R (B) were used in the RT-PCR assays. (C) Constructs (numbers 30-41) containing asAmp with mutated nucleotides (underlined) at the 5′ SS of GTAAGT were co-transfected with plasmid containing ACAT11-1786 (number 27 in Figure 4A) into AC29 cells. Western blot analysis was performed with an anti-Flag antibody. (D, E) Constructs (numbers 42-59, containing ACAT11243-1786 and Ampr) with mutated nucleotides (underlined) at the 3′ SS of CAG were transfected into AC29 cells. Western blot was performed with anti-ACAT1 (D) or anti-Rluc (E) antibodies.
Moreover, detailed mutagenesis experiments showed that the trans-splicing of asAmp transcripts and ACAT1 mRNA was completely interrupted unless a canonical 3′ SS of YAG (Y = C, T) was used at the position of CAG1271-1273(Figure 5D and 5E), confirming the specificity of the 3′ SS. Similarly, serial mutations of the 5′ SS sequence GTAAGT in the plasmid of pVk-3Myc-asAmp (Figure 4A) were generated and the individual plasmids were co-transfected with a Kanr-plasmid pVk-ACAT1-NT-3Flag (Figure 4A, number 27) into AC29 cells. As shown in Figure 5C, the trans-splicing of asAmp and ACAT1 transcripts was completely disrupted unless the sequence of GTRRGN (R = A, G; N = A, G, C, T) was present at the 5′ SS position. Nevertheless, the GTAAGT to GTRRGN mutations at the 5′ SS still led to markedly reduced trans-splicing efficiency (Figure 5C, numbers 34, 37, 40 and 41). Collectively, these data demonstrate that canonical splice-site signals GUAAGU and CAG are utilized in the specific exo-endo trans-splicing of asAmp and ACAT1 transcripts in human cells.
The exo-endo trans-splicing is dependent on the interchromosomal region of the 4.3-kb human ACAT1 chimeric mRNA
We previously reported that the 4.3-kb human ACAT1 chimeric mRNA is most likely produced through an interchromosomal trans-splicing of pre-RNAs transcribed from chromosomes 7 and 110, and that the RNA secondary structures located in the interchromosomal region of the 4.3-kb human ACAT1 chimeric mRNA are required to produce the 56-kDa isoform38. Here, we tested whether the human ACAT1 mRNA interchromosomal region is required for the exo-endo trans-splicing. First, we generated several bicistronic constructs containing the ORF of the Rluc gene as the first cistron and the ORF of the Firefly luciferase (Fluc) gene as the second cistron. These two cistrons were linked by the interchromosomal region (ACAT11243-1396) contained in the 4.3-kb human ACAT1 chimeric mRNA. In certain constructs, a stable hairpin (ΔG = −57 kcal/mol) was introduced upstream (Figure 6A, number 63) or downstream (Figure 6A, number 66) of the first Rluc cistron. We also introduced substitutions of either C to A or C to T at the 3′ SS (CAG1271-1273) within the interchromosomal region (Figure 6A, numbers 61, 62, 64, 65, 67 and 68). After individual transfection of these Ampr-plasmids into AC29 cells, we analyzed their expression by both western blot and RT-PCR analyses. Western blot results demonstrated that the stable hairpin impaired the translation of the first cistron (Figure 6B, bottom panel, numbers 63-65) but not the second one (Figure 6B, top panel, numbers 66-68), suggesting that this interchromosomal region likely contains an internal ribosome entry site as what we have previously reported38. Moreover, only the C to A, not C to T, substitution of the 3′ SS of CAG1271-1273 within the interchromosomal region led to the disappearance of the 68-kDa band corresponding to the larger hybrid protein (Figure 6B, top panel, numbers 61, 64 and 67).
Interchromosomal region of human ACAT1 chimeric mRNA is required for the exo-endo trans-splicing. (A) Schematic representation of bicistrons with or without a stable hairpin. Rluc cistron and Fluc cistron are linked by the ACAT1 interchromosomal region (ACAT11243-1396, derived from both chromosomes 7 and 1). A stable hairpin (ΔG = −57 kcal/mol) is included upstream or downstream of the Rluc cistron. Mutated nucleotides at the 3′ SS are underlined. (B) Western blot of bicistronic proteins using anti-Fluc or anti-Rluc antibodies. (C) RT-PCR analysis of bicistronic RNAs. Specific primer sets A1-F2/FL-R and asAmp-F/FL-R were used in the RT-PCR analysis. All the RT-PCR products were further analyzed by DNA sequencing. Mutated nucleotides at the 3′ SS are indicated by underlines (right and top panel). The trans-splicing junction (dashed line), partial sequences of exon Xa, mini-exon Xb and exon 1 are shown (right and bottom panel). (D) The human ACAT1 interchromosomal region is required for the exo-endo trans-splicing event. Number 69 construct contains the ACAT1 interchromosomal region (ACAT11243-1396). Constructs of number 70 (1-1279+d790) and 71 (u45+1290-1396) contain partial regions of human ACAT1cDNA exclusively derived from chromosome 7 and 1, respectively. RT-PCR assays were performed by using specific primer sets asAmp-F/A1-R2 and asAmp-F/A1-R3 with amplification cycles 25, 29 and 33, synchronously. Reverse primer A1-R3 is complementary to d790. d790 or u45, 790 bp downstream or 45 bp upstream of the exonic sequences. (E) Splicing efficiency of the trans-splicing between asAmp and ACAT1 RNAs in transfected cells was determined by qRT-PCR assays. Spliced and unspliced RNAs were determined with specific primer sets asAmp-F/A1-R2 and A1-F2/A1-R2 as those used in Figure 4G. The percentage of spliced RNAs was calculated by dividing the amount of spliced RNA products by the total RNAs. Means and SD are shown (n = 3). (F) Model of the exo-endo trans-splicing (bold arrow) between the recombinant plasmid-derived asAmp transcript (donor) and the interchromosomal trans-spliced human ACAT1 mRNA (acceptor). The pre-mRNA from either chromosome 7 or 1 is not acceptor of this exo-endo trans-splicing (dashed arrow). The region and orientation of the recombinant plasmid-derived fragment and recombined cryptic promoter are diagramed. Bold characters, AGA and GGC codons at junction site; italic characters, splice-site signals.
These results indicate that the larger 68-kDa hybrid protein (Figure 6B, top panel) was likely produced from trans-spliced mRNAs, and that the substrate mRNAs as the 3′-acceptor must contain the interchromosomal region with CAG as a 3′ SS. Indeed, RT-PCR and DNA sequencing data showed that the ACAT1-Fluc fragment with or without CAG mutations can be individually amplified correctly (Figure 6C, top panel), and that the asAmp transcript can be trans-spliced to the interchromosomal region upstream of Fluc mRNAs with the accurate junction (Figure 6C, right of bottom panel) unless a C to A substitution was introduced at the 3′ SS of CAG (Figure 6C, left of bottom panel, numbers 61, 64 and 67). These results are in perfect accordance with the western blot results shown in Figure 6B (top panel, numbers 61, 64 and 67). Collectively, these data indicate that the interchromosomal region of the 4.3-kb human ACAT1 chimeric mRNA is required for the mRNA trans-splicing process regardless of its linkage with different downstream ORFs such as ACAT1 and Fluc ORF.
Next, we asked whether the asAmp transcript could be directly trans-spliced to pre-RNAs transcribed from chromosome 7 or 1. To test this possibility, we co-transfected AC29 cells with pVk-3Myc-asAmp (Figure 6D, number 26) with plasmids containing the interchromosomal region (Figure 6D, numbers 27 and 69), chromosome 7-derived ACAT1 genomic DNA (Figure 6D, number 70) or chromosome 1-derived ACAT1 genomic DNA (Figure 6D, number 71). RT-PCR analysis showed that although all these plasmids were expressed (Supplementary information, Figure S5), the asAmp transcript was exclusively trans-spliced to the interchromosomal region of ACAT1 chimeric mRNA but not to the pre-RNAs derived from chromosome 7 or 1 (Figure 6D, bottom panel). The splicing efficiency in transfected cells was calculated by qRT-PCR assays. As indicated in Figure 6E, the percentage of spliced RNAs 2 days after the transfection was nearly 10% (Figure 4G).
As diagramed in Figure 6F, we postulate an exo-endo trans-splicing model and conclude that the recombinant plasmid-derived asAmp transcript trans-splices to the 4.3-kb human ACAT1 chimeric mRNA that contains the interchromosomal region derived from chromosomes 7 and 1 (bold arrow), but not to the pre-mRNAs derived from either chromosome 7 or 1 (dashed arrows).
Discussion
We previously reported that the 4.3-kb human ACAT1 chimeric mRNA corresponding to cDNA K1 is presumably produced by an interchromosomal trans-splicing event and give rise to the ACAT1 56-kDa isoform containing an additional region at the N terminus compared with the 50-kDa isoform10,37; this region is now designated as ACAT1-56NT. In the current study, we show that ACAT1-56NT is actually composed of two parts: ACAT1-56eNT, which is comprised of the N-terminal 46 amino acids of asAmp-P88 encoded by the ORF of the asAmp derived from common recombinant Ampr-plasmids, and ACAT1-56uNT, which is comprised of 41 amino acids encoded by the region from GGC1274-1276 to AUG1397-1399, the initiation codon of the ACAT1 50-kDa isoform. Our data obtained in intact-cell and cell-free systems indicate that the human ACAT1 56-kDa isoform is produced by an exo-endo trans-spliced mRNA generated from the plasmid-derived exogenous asAmp transcript and the 4.3-kb human ACAT1 chimeric mRNA (Figure 4). Strikingly, DNA fragments of asAmp with an upstream cryptic promoter, derived from recombinant Ampr-plasmids, and the corresponding exogenous asAmp transcripts have been detected in human blood cells and various cell lines. To our knowledge, this is the first report in mammals of a functional protein produced by the exo-endo trans-splicing of recombinant plasmid-derived exogenous transcripts and endogenous cellular RNAs.
In the 50 blood samples from healthy individuals, the quantity of asAmp DNA was found to be higher in adherent cells than in non-adherent cells (Figure 2). This finding may relate to the health status of individuals in their particular environment and also can be explained by the ability of phagocytes, most of which appear as adherent cells, to obtain the asAmp DNA fragments from phagocytized foreign substances, infectious microbes and other pathogens. In addition, the trans-spliced RNA product and ACAT1 56-kDa isoform do not seem to be present in non-adherent cells (Figure 2F and 2I). This absence may be due to the lack of the 4.3-kb human ACAT1 chimeric mRNA in non-adherent cells, where a prior interchromosomal trans-splicing event may not be induced or the produced ACAT1 chimeric mRNA is quickly degraded44. Our data also show that the exo-endo trans-splicing is dependent on the interchromosomal region of the 4.3-kb human ACAT1 chimeric mRNA (Figure 6), and that the chimeric mRNA is necessary for the production of the ACAT1 56-kDa isoform.
Until now, the detailed mechanism responsible for the presumed interchromosomal trans-splicing events that lead to the production of the 4.3-kb human ACAT1 chimeric mRNA10 has remained obscure. Here, we demonstrated that human ACAT1 56-kDa isoform is the product of an exo-endo trans-splicing event that requires the interchromosomal region of ACAT1 4.3-kb chimeric mRNA. This finding indirectly demonstrates that the interchromosomal trans-splicing must have occurred to produce the 4.3-kb human ACAT1 chimeric mRNA. Also, our findings described here reveal an exo-endo trans-splicing system, in which recombinant plasmid-derived exogenous transcripts are linked with endogenous cellular RNAs in human cells.
Interestingly, an increasing number of observations of chimeric RNAs that are conventionally thought to be formed mainly by trans-splicing, have prompted the proposal of alternative molecular mechanisms for certain chimeric RNAs lacking the canonical splice-site pattern of GU-AG at the splicing junctions16,45. Here, we show that the exo-endo trans-splicing of the asAmp transcripts and the 4.3-kb human ACAT1 chimeric mRNA utilizes canonical splice-site signals (Figure 5), which suggests that this trans-splicing uses regular cellular spliceosome machinery even though the substrates are so unusual. In particular, the interchromosomal region of the ACAT1 chimeric mRNA is necessary for the exo-endo trans-splicing (Figure 6). In addition to the two RNA secondary structures that we have identified38, the potential exonic splicing enhancers in the interchromosomal region of the ACAT1 chimeric mRNA, which are important elements of the “splicing code”46,47, may also be required for the specificity of this exo-endo trans-splicing; these potential sites warrant further investigations.
Antibiotic resistance genes, many of which are found in transposons or integrons, are widely spread throughout the environment48. In the long-term, the antibiotic resistance genes could be mobilized and inserted into eukaryotic genomes by horizontal gene transfer, which may have significant functional implications in both prokaryotic and eukaryotic genome evolution49,50. Here, we demonstrate that DNA fragments of asAmp with an upstream recombined cryptic promoter derived from recombinant Ampr-plasmids and the corresponding exogenous asAmp transcripts exist in higher eukaryotic cells including human blood cells, and their existence was unlikely due to the contamination of plasmids or microbes (Figures 2, 3 and Supplementary information, Figure S2). Furthermore, the quantities of these DNA fragments in human blood cells varied among individual samples. Our findings raise the possibility that Ampr-derived asAmp DNA fragments may be potentially integrated into the genomes of certain higher eukaryotic cells and are expressed as part of cellular RNAs, in part through exo-endo trans-splicing. Interestingly, although DNA fragments of asAmp are broadly identified in eukaryotic cells, amplicons of Kanr are only occasionally observed (Figures 2E and 3A). It may be due to the fact that Ampr-plasmids were used earlier and more extensively than Kanr-plasmids. In the future, it will be interesting to investigate whether and how other exogenous DNA fragments derived from recombinant plasmids affect the human gene expression at both DNA and RNA levels.
Materials and Methods
Cell culture and transfection
All cell lines were maintained in a basal medium as indicated, supplemented with 10% fetal bovine serum (FBS) and antibiotics, in a humid atmosphere of 5% CO2 and 95% air at 37 °C. AC29 cells, the mutant CHO cell line lacking the endogenous ACAT151, were maintained in Ham's F12 medium. HEK293, HEK293T, SV589, HeLa, MCF-7 and A431 cells were maintained in DMEM medium. THP-1, U937 and HL-60 cells were maintained in RPMI 1640 medium. Expression plasmids were transfected into indicated cells using FuGENE 6 transfection reagent (Roche) according to the manufacturer's instructions, and transfected cells were cultured for another 24-36 h before harvesting. Human blood cells were obtained from Shanghai Blood Center (Shanghai, China) and PBMC were separated according to a published procedure52. Adherent and non-adherent cells were separated from the PBMC by incubation in RPMI 1640 medium with 10% FBS for 2 h. The adherent cells were cultured in RPMI 1640 medium supplemented with 7% human type AB serum for another 0-8 days.
PCR and RT-PCR analyses
Genomic DNA was isolated using TIANamp Genomic DNA Kit (TIAGEN Biotech) according to the manufacturer's instructions. Hundred nanograms of DNA was used for PCR or qPCR analysis. Low-molecular-weight plasmid (non-genomic) DNA was extracted following the Hirt extraction method53. Total RNA was isolated using Trizol reagent (Invitrogen), followed by removal of residual DNA using DNase I (Promega). Two microgramsof total RNA was reversely transcribed using oligo (dT)18 and Superscript III RTase (Invitrogen) to obtain relative cDNAs. Twenty-five microlitres PCR reaction consists of 1 μl template DNA or cDNA, 1 U Taq DNA polymerase (TaKaRa) and 0.1 μM of each oligonucleotide primer in nuclease-free water. qPCR was performed by using Brilliant SYBR Green qPCR Master Mix and Mx3005P™ instrument (Stratagene). Products of PCR and RT-PCR were identified by agarose gel electrophoresis, and cycle threshold (Ct) values of qPCR were used to calculate the quantity of each target DNA or cDNA. The relative copy of asAmp DNA per cell was obtained by normalizing to the individual ACAT1 DNA quantity which was set as single copy per cell. All oligonucleotide primers used are listed in Supplementary information, Table S1.
Western blot analysis and 2-D gel electrophoresis
Cells were washed twice, scraped in ice-cold PBS and lysed with RIPA buffer (25 mM Tris-HCl, 150 mM NaCl, 1% NP40, 1% deoxycholate and 0.1% SDS) containing protease inhibitor mixture (Sigma). Protein concentrations were determined using BCA protein assay kit (Bio-Rad). For western blot, each protein sample (50 μg) was separated by SDS-PAGE, transferred to filter and probed with indicated antibodies (Supplementary information, Table S2), then the signals were developed using ECL Western Blot detection reagent (Pierce). Expressed proteins from the AC29 cells transfected with expression plasmid pNTF were purified using ANTI-FLAG M2 Affinity Gel (Sigma), loaded to the IPG strip (pH 3-10 NL) for the isoelectric focusing by IPGphor IEF unit (Pharmacia) and for 2-D analysis of SDS-PAGE. All the experimental procedures for 2-D gel electrophoresis were performed as described54 or following user's manuals.
MALDI-TOF MS analysis and protein N-terminal sequencing
Spots of enriched 26-kDa proteins (P1-P3) separated by 2-D gel electrophoresis were in-gel-digested with trypsin and treated with iodoacetamide. The resultant peptide mixture was desalted and analyzed by MALDI-TOF MS using the instrument Bruker REFLEXTM III MALDI-TOF, located at the Research Center for Proteome Analysis, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. N-terminal amino acids of enriched 26-kDa proteins (P1-P3) were determined by ABI 492cLC protein sequencer (Applied Biosystems) in GeneCore BioTechnologies (Shanghai, China).
Identification of the recombined cryptic promoter by luciferase activity analysis
Upstream regions of asAmp-P88 ORF were amplified from plasmid pcDNA3 by PCR with individual forward primers asAmp-u1000-F (5′-AAACTCGAGAAAGGCGGTAATACGGTTATCCACA-3′), asAmp-u850-F (5′-AAACTCGAGTCGACGCTCAAGTCAGAGGTGG-3′), asAmp-u500-F (5′-AAACTCGAGGCGGTGCTACAGAGTTCTTGAAGTG-3′), asAmp-u275-F (5′-AAACTCGAGGTCTGACGCTCAGTGGAACGAAAA-3′) and common reverse primer asAmp-ATG-R (5′-AAAAAGCTTTGCAGCACTGGGGCCAGATG-3′), and inserted into XhoI and HindIII sites of pGL3-B (Promega) to generate reporter plasmids pasAmp-u1000, pasAmp-u850, pasAmp-u500 and pasAmp-u275, respectively. Cells were washed with ice-cold PBS and lysed with 1× passive lysis buffer (Promega). Cell lysates were subjected to one freeze/thaw cycle at −80 °C and room temperature, respectively, and assayed for luciferase activities using the dual-luciferase reporter assay system (Promega) according to the manufacturer's protocol. Assays were performed using an Auto Lumat BG-P luminometer (MGM Instrument Inc).
In vitro transcription and splicing assay
The linear templates were obtained by PCR using primer sets T7-asAmp (5′-GGATCCTAATACGACTCACTATAGGGATGATACCGCGAGACCCACG-3′)/asAmp-267 (5′-TCTAGATCATGTAACTCGCCTTGA-3′), T7-A1 (5′-GGATCCTAATACGACTCACTATAGGGTAGTTAAATAGCTATATTTAT-3′)/A1-1396 (5′-TCTAGATGTATTGTCTGAGGC-3′) and T7-A1/A1-1786 (5′-TCTAGAATCTAAGAGAGAGCGCC-3′), transcribed using T7 RNA polymerase in the presence of cap analog (Promega) by RiboMAX™ large-scale RNA production systems (Promega). Splicing reactions in cell-free system were performed at 30 °C for 90 min in volumes of 10 μl containing 50% (v/v) HeLa NE(4C Biotec) as previously described42,43. RNAs were purified by phenol/chloroform extraction and ethanol precipitation with glycogen (Sigma), dissolved in RNase-free water and reverse-transcribed using ACAT1 gene-specific primer (5′-GCGGGCACTTCGGC-3′) to obtain the relative cDNAs.
References
Maniatis T, Tasic B . Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 2002; 418:236–243.
Horiuchi T, Aigaki T . Alternative trans-splicing: a novel mode of pre-mRNA processing. Biol Cell 2006; 98:135–140.
Nilsen TW, Graveley BR . Expansion of the eukaryotic proteome by alternative splicing. Nature 2010; 463:457–463.
Nilsen TW . Evolutionary origin of SL-addition trans-splicing: still an enigma. Trends Genet 2001; 17:678–680.
Hastings KE . SL trans-splicing: easy come or easy go? Trends Genet 2005; 21:240–247.
Dorn R, Reuter G, Loewendorf A . Transgene analysis proves mRNA trans-splicing at the complex mod (mdg4) locus in Drosophila. Proc Natl Acad Sci USA 2001; 98:9724–9729.
Horiuchi T, Giniger E, Aigaki T . Alternative trans-splicing of constant and variable exons of a Drosophila axon guidance gene, lola. Genes Dev 2003; 17:2496–2501.
Caudevilla C, Serra D, Miliar A, et al. Natural trans-splicing in carnitine octanoyltransferase pre-mRNAs in rat liver. Proc Natl Acad Sci USA 1998; 95:12185–12190.
Caudevilla C, Codony C, Serra D, et al. Localization of an exonic splicing enhancer responsible for mammalian natural trans-splicing. Nucleic Acids Res 2001; 29:3108–3115.
Li BL, Li XL, Duan ZJ, et al. Human acyl-CoA:cholesterol acyltransferase-1 (ACAT-1) gene organization and evidence that the 4.3-kilobase ACAT-1 mRNA is produced from two different chromosomes. J Biol Chem 1999; 274:11060–11071.
Takahara T, Kanazu SI, Yanagisawa S, Akanuma H . Heterogeneous Sp1 mRNAs in human HepG2 cells include a product of homotypic trans-splicing. J Biol Chem 2000; 275:38067–38072.
Finta C, Zaphiropoulos PG . Intergenic mRNA molecules resulting from trans-splicing. J Biol Chem 2002; 277:5882–5890.
Takahara T, Tasic B, Maniatis T, Akanuma H, Yanagisawa S . Delay in synthesis of the 3′ splice site promotes trans-splicing of the preceding 5′ splice site. Mol Cell 2005; 18:245–251.
Li H, Wang J, Mor G, Sklar J . A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science 2008; 321:1357–1361.
Rickman DS, Pflueger D, Moss B, et al. SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer. Cancer Res 2009; 69:2734–2738.
Herai RH, Yamagishi ME . Detection of human interchromosomal trans-splicing in sequence databanks. Brief Bioinform 2010; 11:198–209.
McManus CJ, Duff MO, Eipper-Mains J, Graveley BR . Global analysis of trans-splicing in Drosophila. Proc Natl Acad Sci USA 2010; 107:12975–12979.
Allen MA, Hillier LW, Waterston RH, Blumenthal T . A global analysis of C. elegans trans-splicing. Genome Res 2011; 21:255–264.
Kannan K, Wang L, Wang J, Ittmann MM, Li W, Yen L . Recurrent chimeric RNAs enriched in human prostate cancer identified by deep sequencing. Proc Natl Acad Sci USA 2011; 108:9172–9177.
Flouriot G, Brand H, Seraphin B, Gannon F . Natural trans-spliced mRNAs are generated from the human estrogen receptor-alpha (hER alpha) gene. J Biol Chem 2002; 277:26244–26251.
Knapp CW, Dolfing J, Ehlert PA, Graham DW . Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ Sci Technol 2010; 44:580–587.
Heuer H, Schmitt H, Smalla K . Antibiotic resistance gene spread due to manure application on agricultural fields. Curr Opin Microbiol 2011; 14:236–243.
Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MO, Dantas G . The shared antibiotic resistome of soil bacteria and human pathogens. Science 2012; 337:1107–1111.
Martínez JL . Antibiotics and antibiotic resistance genes in natural environments. Science 2008; 321:365–367.
Salyers AA, Gupta A, Wang Y . Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol 2004; 12:412–416.
Cohen SN, Chang AC, Boyer HW, Helling RB . Construction of biologically functional bacterial plasmids in vitro. Proc Natl Acad Sci USA 1973; 70:3240–3244.
Lobban PE, Kaiser AD . Enzymatic end-to end joining of DNA molecules. J Mol Biol 1973; 78:453–471.
Peter W, Alberts B, Johnson AS, Lewis J, Raff MC, Roberts K, eds. Molecular Biology of the Cell, 5th Edition. New York: Garland Science, 2008.
Funke T, Han H, Healy-Fried ML, Fischer M, Schönbrunn E . Molecular basis for the herbicide resistance of Roundup Ready crops. Proc Natl Acad Sci USA 2006; 103:13010–13015.
Hanahan D . Heritable formation of pancreatic beta-cell tumours in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes. Nature 1985; 315:115–122.
Chang CC, Sun J, Chang TY . Membrane-bound O-acyltransferases (MBOATs). Front Biol 2011; 6:177–182.
Chang TY, Li BL, Chang CC, Urano Y . Acyl-coenzyme A:cholesterol acyltransferases. Am J Physiol Endocrinol Metab 2009; 297:E1–E9.
Chang TY, Chang CC, Cheng D . Acyl-coenzyme A:cholesterol acyltransferase. Annu Rev Biochem 1997; 66:613–638.
Yu C, Chen J, Lin S, Liu J, Chang CC, Chang TY . Human acyl-CoA:cholesterol acyltransferase-1 is a homotetrameric enzyme in intact cells and in vitro. J Biol Chem 1999; 274:36139–36145.
Guo ZY, Lin S, Heinen JA, Chang CC, Chang TY . The active site His-460 of human acyl-coenzyme A:cholesterol acyltransferase 1 resides in a hitherto undisclosed transmembrane domain. J Biol Chem 2005; 280:37814–37826.
Chang CC, Huh HY, Cadigan KM, Chang TY . Molecular cloning and functional expression of human acyl-coenzyme A:cholesterol acyltransferase cDNA in mutant Chinese hamster ovary cells. J Biol Chem 1993; 268:20747–20755.
Yang L, Lee O, Chen J, et al. Human acyl-coenzyme A:cholesterol acyltransferase 1 (acat1) sequences located in two different chromosomes (7 and 1) are required to produce a novel ACAT1 isoenzyme with additional sequence at the N terminus. J Biol Chem 2004; 279:46253–46262.
Chen J, Zhao XN, Yang L, et al. RNA secondary structures located in the interchromosomal region of human ACAT1 chimeric mRNA are required to produce the 56 kDa isoform. Cell Res 2008; 18:921–936.
Yang L, Chen J, Chang CC, et al. A stable upstream stem-loop structure enhances selection of the first 5′-ORF-AUG as a main start codon for translation initiation of human ACAT1 mRNA. Acta Biochim Biophys Sin 2004; 36:259–268.
Santos SR, Ochman H . Identification and phylogenetic sorting of bacterial lineages with universally conserved genes and proteins. Environ Microbiol 2004; 6:754–759.
Zhang Z, Schwartz S, Wagner L, Miller W . A greedy algorithm for aligning DNA sequences. J Comput Biol 2000; 7:203–214.
Anderson K, Moore MJ . Bimolecular exon ligation by the human spliceosome. Science 1997; 276:1712–1716.
Reichert V, Moore MJ . Better conditions for mammalian in vitro splicing provided by acetate and glutamate as potassium counterions. Nucleic Acids Res 2000; 28:416–423.
Zhao XN, Chen J, Lei L, et al. The optional long 5′-untranslated region of human ACAT1 mRNAs impairs the production of ACAT1 protein by promoting its mRNA decay. Acta Biochim Biophys Sin 2009; 41:30–41.
Li X, Zhao L, Jiang H, Wang W . Short homologous sequences are strongly associated with the generation of chimeric RNAs in eukaryotes. J Mol Evol 2008; 68:56–65.
Fu XD . Towards a splicing code. Cell 2004; 119:736–738.
Wang J, Smith PJ, Krainer AR, Zhang MQ . Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes. Nucleic Acids Res 2005; 33:5053–5062.
Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J . Call of the wild: antibiotic resistance genes in natural environments. Nature Rev Microbiol 2010; 8:251–259.
Salzberg SL, White O, Peterson J, Eisen JA . Microbial genes in the human genome: lateral transfer or gene loss. Science 2001; 292:1903–1906.
Keeling PJ, Palmer JD . Horizontal gene transfer in eukaryotic evolution. Nature Rev Genet 2008; 9:605–618.
Cadigan KM, Heider JG, Chang TY . Isolation and characterization of Chinese hamster ovary cell mutants deficient in acylcoenzyme A:cholesterol acyltransferase activity. J Biol Chem 1988; 263:274–282.
Ouchi N, Kihara S, Arita Y, et al. Adipocyte-derived plasma protein, adiponectin, suppresses lipid accumulation and class a scavenger receptor expression in human monocyte-derived macrophages. Circulation 2001; 103:1057–1063.
Hirt B . Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol 1967; 26:365–369.
Garrels JI . Quantitative two-dimensional gel electrophoresis of proteins. Methods Enzymol 1983; 100:411–423.
Acknowledgements
We thank our colleagues Xin-Ying Yang, Lei Lei, Xi-Han Hu and Lei Qian for technical assistances; Prof Guo-Liang Xu for advice and generous sharing of HeLa cell nuclear extracts; Prof Ling-Ling Chen, Li Yang and Jing-Yi Hui for helpful discussions. This work was supported by grants (2011CB910900 and 2009CB919000) from the Ministry of Science and Technology of China to Bo-Liang Li and Bao-Liang Song, and a grant (HL 60306) from the National Institutes of Health to Ta-Yuan Chang and Catherine Chang.
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
Analysis of purified 26-kDa proteins by MALDI-TOF MS. (PDF 45 kb)
Supplementary information, Figure S2
Analysis of asAmp DNA and asAmp transcript in human peripheral blood mononuclear cells (PBMC). (PDF 98 kb)
Supplementary information, Figure S3
Identification of DNA fragments of asAmp with the upstream recombined cryptic promoter. (PDF 200 kb)
Supplementary information, Figure S4
Preparation of 5′-donor (D1) and 3′-acceptors (A1 and A2). (PDF 25 kb)
Supplementary information, Figure S5
RT-PCR analysis of asAmp transcripts, human ACAT1 chimeric mRNAs and RNAs derived from chromosome 7 or 1. (PDF 77 kb)
Supplementary information, Table S1
Oligonucleotide primers (PDF 31 kb)
Supplementary information, Table S2
Antibody reagents (PDF 12 kb)
Supplementary information, Table S3
Primers for construction of plasmids (PDF 14 kb)
Supplementary information, Data S1
Materials and Methods (PDF 73 kb)
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0
About this article
Cite this article
Hu, GJ., Chen, J., Zhao, XN. et al. Production of ACAT1 56-kDa isoform in human cells via trans-splicing involving the ampicillin resistance gene. Cell Res 23, 1007–1024 (2013). https://doi.org/10.1038/cr.2013.86
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2013.86
Keywords
This article is cited by
-
Antibiotic resistomes in water supply reservoirs sediments of central China: main biotic drivers and distribution pattern
Environmental Science and Pollution Research (2022)
-
Trans-spliced long non-coding RNA: an emerging regulator of pluripotency
Cellular and Molecular Life Sciences (2018)
-
Exo-endo trans splicing: a new way to link
Cell Research (2013)