Abstract
In response to nutrient stress, cells start an autophagy program that can lead to adaptation or death. The mechanisms underlying the signaling from starvation to the initiation of autophagy are not fully understood. In the current study we show that the absence or inactivation of PARP-1 strongly delays starvation-induced autophagy. We have found that DNA damage is an early event of starvation-induced autophagy as measured by γ-H2AX accumulation and comet assay, with PARP-1 knockout cells displaying a reduction in both parameters. During starvation, ROS-induced DNA damage activates PARP-1, leading to ATP depletion (an early event after nutrient deprivation). The absence of PARP-1 blunted AMPK activation and prevented the complete loss of mTOR activity, leading to a delay in autophagy. PARP-1 depletion favors apoptosis in starved cells, suggesting a pro-survival role of autophagy and PARP-1 activation after nutrient deprivation. In vivo results show that neonates of PARP-1 mutant mice subjected to acute starvation, also display deficient liver autophagy, implying a physiological role for PARP-1 in starvation-induced autophagy. Thus, the PARP signaling pathway is a key regulator of the initial steps of autophagy commitment following starvation.
Similar content being viewed by others
Introduction
Nutrient starvation alarms eukaryotic cells to adjust metabolism to survive. An early response of the cellular metabolic adjustments involves inhibition of growth and induction of macroautophagy (referred to as autophagy) to optimize the usage of limited energy supplies. Autophagy, as a cellular process mobilizing intracellular nutrient resources, plays an important role in contributing to survival during these growth-unfavorable conditions. It is a highly conserved self-eating process in which intracellular membrane structures engulf a portion of cytoplasmic organelles for lysosomal degradation. Eukaryotic cells have developed a mechanism through which autophagy induction is tightly coupled to the regulation of cell growth. Disruption of autophagic pathways is associated with multiple disease states, including neurodegenerative diseases, cancer, infection, and several types of myopathy 1. Autophagy is also a major mechanism by which starving cells reallocate nutrients from unnecessary to more essential processes 1. During autophagy, a cytosolic form of light chain 3 (LC3; LC3-I) is cleaved and then conjugated to phosphatidylethanolamine to form the LC3-phosphatidylethanolamine conjugate (LC3-II), which is recruited to autophagosomal membranes 2. Poly(ADP-ribose) (PAR) polymerase (PARP) enzymes catalyze the conversion of NAD+ to polymers of PAR 3. Although its role in the DNA damage response has long been recognized, recent works indicate that PAR itself acts to directly induce cell death through stimulation of apoptosis-inducing factor (AIF) release 4, 5.
A recent study from our group has also implicated PARP-1 in autophagy induced by DNA damage and oxidative stress 6. There are, however, important issues that remain unresolved, such as the involvement of PARP signaling in a physiologic model of autophagy as is the case for nutrient deprivation and the connection of PARP activation with the autophagic components. Among the numerous factors involved in the regulation of autophagy and growth, mTOR (target of rapamycin) is a key component that coordinately regulates the balance between growth and autophagy in response to physiological conditions and environmental stress.
In the current study we have found that starvation-induced autophagy results in reactive oxygen species (ROS) production, DNA damage (as measured by comet assay and γ-H2AX accumulation) and PARP-1 activation, leading to the inhibition of mTOR. Moreover, parp-1−/− neonates display a deficient autophagy response following acute starvation. Altogether these results place PARP-1 activation and PAR formation as key players in the decision of the cell to engage autophagy.
Results
The absence or inhibition of PARP-1 delays starvation-induced autophagy
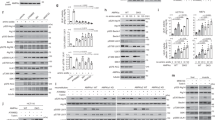
Starvation or nutrient deprivation is a physiological cellular stress to induce autophagy in eukaryotic cells. To study the role of PARP-1 in starvation-induced autophagy, we transiently transfected parp-1+/+ and parp-1−/− mouse embryonic fibroblasts (MEFs) with GFP-LC3 and starved these cells with HANK buffer for 1, 2 and 4.5 h. The percentage of cells with punctate pattern of GFP-LC3 was counted by fluorescence microscopy. In the non-starved cells, GFP-LC3 was diffusely distributed in the cytosol and nucleus, but after treatment with HANK buffer there was a punctate pattern, indicative of an accumulation of autophagosomes (Figure 1A). The number of GFP-LC3 vesicles was higher in parp-1+/+ MEFs, with approximately 20 vesicles/cell in parp-1+/+ MEFs and 8-9 vesicles/cell in parp-1−/− MEFs at 2 h of starvation (Supplementary information, Figure S1). At different times of starvation, a decreased number of cells with GFP-LC3 punctate pattern was observed in parp-1−/− MEFs (Figure 1A). Rapamycin, an inhibitor of mTORC1, was used as a positive control for autophagy induction; parp-1−/− cells were also less sensitive to rapamycin-induced autophagy than wild-type (WT) cells (Figure 1A). The conversion of LC3-I to LC3-II through proteolytic cleavage and lipidation is a hallmark of mammalian autophagy. We measured the LC3 conversion during starvation in parp-1+/+ and parp-1−/− cells, and found that it was decreased in parp-1−/− MEFs (Figure 1B). These data indicate a pronounced delay of autophagy in the absence of PARP-1.
Starvation-induced autophagy is delayed in PARP-1 knockout cells. (A) parp-1+/+ and parp-1−/− MEFs were transfected with GFP-LC3; 24 h following transfection, cells were starved with HANK buffer at 1, 2 and 4.5 h; percentages of LC3 conversion are shown. Treatment with 100 nM rapamycin for 4 h was used as positive control of autophagosome accumulation. The pictures in the right panel show representative images with the subcellular distribution of the autophagic vesicle marker LC3. *P < 0.05 comparing between starved parp-1+/+ and parp-1−/− MEFs. (B) Immunoblot analysis of endogenous LC3 conversion in control and starved cells at different times of starvation. Treatment with rapamycin was used as positive control of LC3 conversion and β-actin as loading control. Similar results were obtained in three independent experiments. *P < 0.05 comparing between starved parp-1+/+ and parp-1−/− MEFs. #P < 0.05 comparing between rapamycin-treated parp-1−/− MEFs and non-starved control parp-1−/− MEFs. (C) Effect of 2 mM 3-MA on autophagy of parp-1+/+ and parp-1−/− MEFs during starvation. 3-MA was added 1.5 h before HANK buffer as pre-treatment and maintained during the starvation in both cells lines to slow down autophagy. **P < 0.01 comparing between starved parp-1+/+ and parp-1−/− MEFs. #P < 0.05 comparing between starved parp-1+/+ MEFs and 3-MA-treated parp-1+/+ MEFs. (D) Effect of ATG7 silencing on starvation-induced autophagy. parp-1+/+ and parp-1−/− MEFs were transfected with ATG7 siRNA (60 nM) and 48 h later they were transfected with GFP-LC3. 24 h later cells were starved with HANK buffer for 30 min, 1 h and 2 h. SIMA or non-specific siRNA was used as negative control, using the same protocol as for siRNA transfection. The right panel show the siRNA-mediated suppression of ATG7 expression in MEFs 3T3 48 h after transfection. GAPDH was used as loading control. *P < 0.05 comparing between starved parp-1+/+ and parp-1−/− MEFs. #P < 0.05 comparing between starved parp-1+/+ MEFs and ATG7-knockdown parp-1+/+ MEFs. Western blot quantification: ***P < 0.001 comparing between ATG7-knockdown parp-1+/+ MEFs and control parp-1+/+ MEFs. ##P < 0.01 comparing between ATG7-knockdown parp-1−/− MEFs and control parp-1−/− MEFs. In A, C and D, at least 250 cells were counted under a Zeiss fluorescent microscope in both cell lines in three independent experiments.
To further evaluate autophagy in this model, we used a chemical inhibitor of autophagy 3-Methyladenine (3-MA), an inhibitor of class III phosphatidylinositol 3-kinase 7, as well as the siRNA-based knockdown of an essential autophagy protein, Atg7. Treatment with 3-MA or siRNA of Atg7 led to a significant reduction in the number of cells with GFP-LC3 punctate pattern in parp-1+/+ MEFs after 2 h of starvation (Figure 1C and 1D). In starved parp-1−/− MEFs, 3-MA or Atg7 siRNA treatment completely prevented autophagy (Figure 1C, 1D and Supplementary information, Figure S2). These data suggest that the translocation of GFP-LC3 observed in parp-1+/+ MEFs upon starvation is due to autophagy and reflects the functional role of autophagy during starvation. The absence of PARP-1 synergizes with 3-MA or ATG7 siRNA to suppress autophagy during starvation (Figure 1C and 1D). Lysosome fusion with autophagosomes was not affected in parp-1−/− cells. Treatment with chloroquine to inhibit lysosome fusion resulted in a similar accumulation of LC3 vesicles in parp-1+/+ and parp-1−/− cells (Supplementary information, Figure S3A).
To further evaluate the role of PARP-1 in starvation-induced autophagy, we tested the effect of the PARP-1 inhibitor DPQ and siRNA-based depletion of PARP-1 on the levels of autophagy in parp-1+/+ MEFs. Cells were transfected with GFP-LC3 and starved with HANK buffer for different time periods. Inhibition of PARP-1 with 40 μM DPQ reduced the number of cells with a typical GFP-LC3 punctate pattern in starved parp-1+/+ MEFs, but had no effect in parp-1−/− MEFs (Figure 2A). Similar results were also obtained using two other different PARP inhibitors, PJ34 and olaparib (Supplementary information, Figure S3B). PARP-1 silencing induced a reduction in the number of autophagic cells after 2 h of starvation (Figure 2B), similar to that in parp-1−/− cells (Figure 2A). Further, PARP-1 knockdown reduced the conversion of endogenous LC3 during starvation (Figure 2C). The non-specific siRNA had no effect on the levels of autophagy. These data suggest that PARP-1 and PARP activation play an active role in the commitment to autophagy in situations of nutrient deprivation. To corroborate this finding, we reconstituted PARP-1 expression in parp-1−/− MEFs with pBC-PARP-1 cDNA (Figure 2D and Supplementary information, Figure S4) and we co-transfected these cells with GFP-LC3. Cells transfected with the empty pBC vector were used as negative control. The reconstitution of PARP-1 in parp-1−/− MEFs increased the number of autophagic cells upon starvation compared to parp-1−/− MEFs transfected with the empty vector (Figure 2D and Supplementary information, Figure S4). These data suggest that PARP-1 participates in the commitment of starvation-induced autophagy and may be indirectly involved in the formation of autophagosomes, since its inhibition or silencing leads to a delay in the autophagic response, including LC3 conversion and autophagosome formation. Indeed, autophagy was delayed but not abrogated after PARP-1 ablation, since increasing starvation time to 8 h resulted in an equivalent autophagic rate between WT and knockout cells (data not shown).
PARP-1 inhibition or silencing interferes with starvation-induced autophagy. (A) Effect of the inhibition of PARP-1 with DPQ on starvation-induced autophagy. parp-1+/+ and parp-1−/− MEFs were transfected with GFP-LC3; 24 h later, cells were pre-treated with 40 μM DPQ for 1.5 h. During the different starvation time periods, 40 μM DPQ was present in HANK buffer to maintain PARP-1 inhibition. Percentages of cells with LC3 conversion are shown. *P < 0.05 comparing between DPQ-treated starved parp-1+/+ MEFs and starved parp-1+/+ MEFs. (B) Effect of PARP-1 silencing on starvation-induced autophagy. parp-1+/+ MEFs were transfected with murine PARP-1 siRNA (50 nM). 48 h later cells were transfected with GFP-LC3 and 24 h later were staved for 2 h. The percentage of cells with the typical GFP-LC3 punctate pattern was compared with the percentage in non-silencing parp-1+/+ MEFs starved for the same time period. SIMA or non-specific siRNA (50 nM) was used as negative control, using the same protocol as for siRNA transfection. The right panel shows the levels of PARP-1 silencing 48 h after the transfection. α-Tubulin was used as loading control. *P < 0.05 comparing between starved parp-1+/+ MEFs and PARP-1-knockdown starved parp-1+/+ MEFs. (C) Western blot analysis of the effect of PARP-1 silencing on endogenous LC3 conversion in parp-1+/+ MEFs starved for 2 h with HANK buffer. α-Tubulin was used as loading control. Similar results were obtained in three independent experiments. **P < 0.01 comparing between starved parp-1+/+ MEFs and PARP-1-knockdown starved parp-1+/+ MEFs. (D) Reconstitution of PARP-1 in parp-1−/− MEFs and effect on starvation-induced autophagy. parp-1−/− MEFs were transfected with pBC-PARP-1 to transiently reconstitute PARP-1 and 24 h later cells were transfected with GFP-LC3; 24 h after transfection cells were starved with HANK buffer for 30 min and 2 h. pBC empty plasmid was used as negative control. The right panel shows the expression level of PARP-1 24 h after reconstitution in parp-1−/− MEFs and compared with the expression levels of PARP-1 in WT and knockout cells. α-Tubulin was used as loading control. *P < 0.05 comparing between starved parp-1−/− MEFs and PARP-1-restored parp-1−/− MEFs. In A, B and D, at least 250 nuclei were counted under a Zeiss fluorescent microscope in three independent experiments.
Starvation induces ROS production, DNA damage and activation of PARP-1
PARP-1 is a nuclear enzyme activated by DNA damage; following genotoxic stress PARP-1 synthesizes a branched polymer of poly(ADP-ribose) or PAR that participates in the regulation of the nuclear homeostasis 3, 6, 8. Many different cellular insults that cause DNA damage activate PARP-1 and induce PARP-1-dependent cell death. During starvation the production of ROS plays an important role in triggering autophagy 9, 10. We hypothesized that ROS production during starvation could induce activation of PARP-1 and play an important role in the regulation of PARP-1-dependent autophagy.
Starvation indeed induced activation of PARP-1 in parp-1+/+ MEFs, as measured by PAR synthesis (Figure 3A). The PARP inhibitor PJ34 blocked starvation-induced PAR synthesis, and H2O2 was used as a positive control for activation of PARP-1 (Figure 3A and Supplementary information, Figure S5).
PAR synthesis and DNA damage during starvation-induced autophagy. (A) Western blot analysis of PAR formation during starvation. parp-1+/+ MEFs were starved for 30 min, 1 h and 2 h with HANK buffer. Whole cell extracts were subjected to PAGE and PAR was measured with a specific antibody. Where indicated, cells were pre-treated with PJ34, a PARP-1 inhibitor, for 1.5 h and PJ34 was maintained during the nutrient deprivation. Treatment with 10 mM H2O2 for 10 min was used as a positive control of PARP-1 activation and PAR synthesis. α-Tubulin was used as loading control. (B) ROS production during starvation. parp-1+/+ and parp-1−/− MEFs were subjected to short times of nutrient deprivation with HANK buffer in the presence of the DCFDA probe (8 mg/ml), specific to measure ROS with a flow cytometer. Figure shows DCFDA fluorescence obtained as arbitrary units in triplicate in three independent experiments. *P < 0.05 comparing between starved parp-1+/+ and parp-1−/− MEFs. (C) COMET assay during starvation. parp-1+/+ and parp-1−/− MEFs were starved with HANK buffer for 15, 30 and 60 min and then were treated according to the manufacturer's instruction. Tail moment of 90 nuclei per condition in three independent experiments were analyzed by the specific software CASP (left panel). In the right panel images of the COMETs extracted from the software of each cell line for the different times of starvation are shown. **P < 0.01 comparing between starved parp-1+/+ and parp-1−/− MEFs. (D) Histone γ-H2AX phosphorylation during starvation. parp-1+/+ and parp-1−/− MEFs were subjected to starvation for the indicated times. Total extract were obtained and the levels of phospho-γ-H2AX were measured by immunoblotting. Similar results were obtained in three independent experiments. α-Tubulin was used as loading control. *P < 0.05, **P < 0.01 and ***P < 0.001 comparing between starved and non-starved parp-1+/+ MEFs. #P < 0.05, ##P < 0.01 comparing between starved and non-starved parp-1−/− MEFs. (E) Treatment with the antioxidant NAC results in a significant delay in mTOR inactivation during starvation-induced autophagy. LC3 western blot quantification: **P < 0.01, ***P < 0.001 comparing between starved parp-1+/+ MEFs and NAC-treated starved parp-1+/+ MEFs. Phospho-γ-H2Ax western blot quantification: **P < 0.01 comparing between starved parp-1+/+ MEFs and NAC-treated starved parp-1+/+ MEFs.
To demonstrate the production of ROS during starvation, we used 2′,7′-dichlorofluorescein diacetate (DCFDA) as a probe to measure ROS (in particular, this probe detects H2O2) in parp-1+/+ and parp-1−/− MEFs by flow cytometry 11. PARP-1-deficient cells displayed a reduced production of ROS even at very early time points following starvation (Figure 3B). This finding is consistent with previous results showing a reduced ROS production in lymphocytes challenged with exogenous oxidative stress and treated with PARP inhibitors 12. Assuming that ROS synthesis and their nuclear diffusion to induce DNA damage are very fast, we chose 30 min of starvation as the time point to measure DNA damage. COMET assay showed that 30 min after starvation, DNA damage was much more pronounced in parp-1+/+ MEFs (Figure 3C); the tail moment (TM) of the comets is much higher in parp-1+/+ cells, and almost full repair is achieved after 1 h, indicating that the DNA repair machinery is active (Figure 3C). By contrast, in parp-1−/− cells the level of DNA damage is clearly reduced at 30 min, but the DNA repair machinery is not as efficient as in WT cells, resulting in a residual level of damage after 60 min of starvation (Figure 3C).
At the same time, parp-1+/+ MEFs had higher levels of phosphorylation of γ-H2AX, suggesting an increased number of DNA lesions due to the boost in ROS production (Figure 3D). γ-H2AX signal peaks at 1 h of starvation in parp-1+/+ MEFs while in PARP-1-deficient cells γ-H2AX continues to accumulate, consistent with a less efficient repair as implicated by the COMET assay. Furthermore, indirect immunofluoresence analysis revealed that the number of parp-1+/+ cells with positive staining for γ-H2AX after 1 h of starvation was elevated compared to parp-1−/− cells (Supplementary information, Figure S6).
To further confirm the implication of ROS in the initiation of autophagy, we used the antioxidant n-acetylcysteine (NAC). Cells exposed to this compound showed a decrease in γ-H2AX accumulation and LC3II lipidation, indicating that ROS generation is key in triggering DNA damage and subsequent autophagy (Figure 3E).
Together, these data indicate that during starvation there is an important production of ROS in parp-1+/+ MEFs and these ROS induce DNA damage and PARP-1 activation, leading to PAR synthesis and triggering the initiation of autophagy associated to nutrient deprivation. Although parp-1−/− cells also produce ROS during starvation, this production does not lead to massive DNA damage and PARP-1 activation; consequently these cells display an impaired starvation-induced autophagy.
Lack of PARP-1 reduces ATP depletion, AMPK activation and mTOR inhibition during starvation-induced autophagy
Energy depletion, measured as an imbalance of AMP/ATP (adenosine 5′-triphosphate) ratio, is the main signal sensed by AMPK to induce autophagy. To investigate whether PARP-1 is implicated in AMPK-dependent autophagy, we measured the levels of ATP in parp-1+/+ MEFs and parp-1−/− MEFs after different times of starvation. The levels of ATP in parp-1+/+ MEFs after 60 min of starvation decreased to less than 50% of the initial level, while in parp-1−/− MEFs ATP levels decreased significantly more slowly (Figure 4A). Treatment with 3-MA during starvation blocked ATP depletion in parp-1+/+ cells, indicating that this energy drop was due to autophagy induced by nutrient deprivation (Figure 4A). 3-MA also prevented ATP depletion in parp-1−/− cells, which have delayed autophagy. The depletion of ATP level corresponded with a sustained activation, through phosphorylation, of AMPK in parp-1+/+ MEFs (Figure 4B). This activation was strongly inhibited in parp-1−/− MEFs (Figure 4B).
PARP-1 modulates AMPK activation and mTOR inhibition during starvation-induced autophagy. (A) Effect of nutrient starvation on ATP levels. parp-1+/+ and parp-1−/− MEFs were starved with HANK buffer for 15, 30 and 60 min. Concentrations of ATP were normalized with total proteins in each sample and compared to the control (100%). 2 mM 3-MA was added 1.5 h before nutrient deprivation and was maintained during the experiment in order to inhibit autophagy. Error bars represent SE of the mean (SEM) of five independent experiments. (B) Immunoblot analysis of AMPK activation in control and starved cells at different times of starvation. The levels of phospho-AMPK in whole cell extracts were measured with a specific antibody in each cell lines after nutrient deprivation. Total AMPK was used to normalize the non-phosphorylated protein and β-actin as loading control. Similar results were obtained in three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 comparing between starved parp-1+/+ MEFs and non-starved control parp-1+/+ MEFs. #P < 0.05 comparing between starved parp-1−/− MEFs and non-starved control parp-1−/− 3T3 MEFs. (C) Immunoblot analysis of mTOR inhibition during starvation. The levels of phosphorylation of the mTOR substrate, p70S6 kinase, were measured by western blotting in whole cell extracts of parp-1+/+ and parp-1−/− MEFs after different times of starvation. Treatment with concanamycin A (2 nM) for 4 h was used as control of mTOR-independent autophagy while treatment with rapamycin (100 nM) for 6 h was used as control of mTOR-dependent autophagy. Total p70S6 kinase was used to normalize the non-phosphorylated protein and β-actin as loading control. Similar results were obtained in two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 comparing between starved parp-1+/+ MEFs and non-starved control parp-1+/+ MEFs. ##P < 0.01 comparing between starved parp-1−/− MEFs and non-starved control parp-1−/− 3T3 MEFs. (D) Treatment with the antioxidant NAC significantly delays starvation-induced loss of mTOR activation measured as phospho-p70S6 kinase. **P < 0.01 comparing between starved parp-1+/+ MEFs and non-starved control parp-1+/+ MEFs. #P < 0.05 comparing between NAC-treated and starved parp-1+/+ MEFs and NAC-treated and non-starved control parp-1+/+ 3T3 MEFs. (E) Induction of autophagy in MCF7-GFP-LC3 during starvation. Treatment with rapamycin was the positive control of autophagy. *P < 0.05 comparing between starved MCF7-GFP-LC3 and 3-MA-treated starved MCF7-GFP-LC3. (F) PARP-1 knockdown prevents autophagy-induced mTOR inhibition in a tumor cell model. Left panel: MCF7-GFP-LC3 cells were starved for different times with HANK buffer and the levels of phospho-p70S6 kinase were measured by western blotting. Concanamycin A and rapamycin were different controls of mTOR activation. p70S6 kinase and GAPDH were used to normalize protein loading. The results were obtained in 3 independent experiments. Right panel: MCF7-GFP-LC3 cells were either non-transfected, transfected with a scrambled (SIMA) siRNA or with PARP-1 siRNA (60 nM) and 48 h after transfection, cells were starved with HANK buffer; the levels of phospho-p70S6 kinase were measured by western blot. Concanamycin A and rapamycin were used as controls of mTOR activation. Total p70S6 kinase and GAPDH were used to normalize protein loading. Similar results were obtained in 3 independent experiments. *P < 0.05, **P < 0.01 comparing between starved MCF7-GFP-LC3 and non-starved control MCF7-GFP-LC3. #P < 0.05, ##P < 0.01 comparing between PARP-1-konckdown starved MCF7-GFP-LC3 and non-starved control MCF7-GFP-LC3.
mTOR is a serine/threonine protein kinase that regulates cell growth, cell proliferation, cell motility, cell survival, protein synthesis, and transcription 13. mTOR is also a sensor of the cellular energy status. For this action it is regulated by the kinase AMPK, an important activator of autophagy 14, 15. We have thus evaluated the status of mTOR (which turns off autophagy when it is activated) by determining the phosphorylation of its substrate p70S6 kinase. Shortly after starvation (30 min) parp-1+/+ cells attained a complete inhibition of mTOR (Figure 4C), indicating that these cells were engaged in autophagy, while in parp-1−/− cells mTOR was only partially inhibited even after 1 and 2 h of nutrient starvation; 4 h after starvation mTOR activation started to recover in the absence of PARP-1 (Figure 4C). Treatment with antioxidant NAC retarded starvation-induced mTOR inactivation (Figure 4D). These data suggest that PARP-1 may control mTOR activity during starvation and functions as a positive regulator of autophagy when cellular energy declines.
To further explore this, we analyzed the role of PARP-1 in mTOR activation regulation during starvation in a tumor cell model. In cancer cells the energy balance is critical to adapt the cell to the tumor microenvironment, which often features low nutrient and oxygen availability 16, 17. We have performed assays with human breast cancer cell line stably transfected with GFP-LC3, MCF7-GFP-LC3. During starvation these cells showed endogenous LC3 translocation and the typical punctate pattern of GFP-LC3 under fluorescence microscope (Figure 4E). MCF7-GFP-LC3 cells were used to determine the level of mTOR activation during starvation in the presence or absence of PARP-1 (Figure 4F). In these cells, mTOR is rapidly inhibited after starvation; 15 min after nutrient deprivation the phosphorylation levels of p70S6 kinase decreased drastically and the cells had entered into autophagy. Treatment with 3-MA retarded starvation-induced mTOR inactivation and downregulated AMPK (Supplementary information, Figure S7). In MCF7-GFP-LC3 depleted of PARP-1 using siRNA (Figure 4F), mTOR inhibition was significantly delayed. The delayed inhibition of mTOR activity in PARP-1 knockout cells and after PARP-1 silencing suggests that disabling of PARP-1 regulates autophagy by preventing complete mTOR inactivation.
To further confirm the implication of efficient DNA damage repair in the initiation of autophagy, we used HT144 melanoma cells (an ATM-deficient cell line) that are unable to properly repair γ-irradiation-induced DNA damage (data not shown). These cells displayed very high levels of DNA damage under basal conditions as shown by the elevated constitutive γ-H2AX levels (Supplementary information, Figure S8A). G361 cells (ATM WT melanoma cells) accumulated DNA damage following starvation and inhibition of mTOR/p70S6K was achieved after 15 min. mTOR/p70S6K activity also decreased very rapidly in ATM mutant cells, but a residual activation was still detected after 30 min of starvation (Supplementary information, Figure S7A). The execution of autophagy determined as LC3 processing (Supplementary information, Figure S8B) and quantification of LC3 punctate cells (Supplementary information, Figure S8C) was delayed in ATM mutant cells, further supporting the mechanistic implication of DNA damage repair in the cell's ability to engage autophagy.
Impairment of autophagy by disabling of PARP-1 leads to increased apoptotic cell death
Autophagy is an adaptation and survival pathway under adverse conditions, but if these conditions are maintained for a long time, excessive autophagy can lead to cell death, often called autophagic cell death (ACD) 18. ACD is morphologically defined as a type of cell death that occurs in the absence of chromatin condensation, but it is accompanied by massive autophagic vacuoles in the cytoplasm 19. In contrast with the typical apoptotic cell death, in ACD there is no nuclear fragmentation, plasma membrane blebbing, caspase activation or engulfment by phagocytes in vivo 20. Cells undergoing autophagy under adverse conditions can recover to their optimum physiological state after changing of the surrounding conditions. However, cells with inhibited autophagy or cells with disrupted mitochondrial transmembrane potential, would die even if optimal conditions are recovered 21. To determine what is the predominant cell death process during starvation-induced autophagy upon ablation/inactivation of PARP-1, we examined both ACD and apoptotic cell death features after prolonged time of starvation. Time course analysis of total cell death following starvation showed accelerated cell death in the absence of PARP-1 as determined by propidium iodide (PI) incorporation (Figure 5A). PARP-1 inhibition with PJ34 or PARP-1 knockdown gave similar results of increased cell death after nutrient deprivation (Figure 5A). Thus, the absence of PARP-1 during starvation accelerates cell death. These results were confirmed by other methods of cell death assays such as trypan blue dye exclusion staining and MTT cell survival assay and with a different PARP-1 inhibitor DPQ (data not shown).
Pro-survival autophagy is switched to apoptosis after PARP-1 ablation. (A) Effect of PARP inhibition and PARP-1 silencing on cell death during autophagy induced by nutrient deprivation. parp-1+/+ MEFs were transfected with PARP-1 siRNA (60 nM) and 48 h after transfection, cells were pre-treated with or without PJ34 (10 μM). Cells were starved for 8 h with HANK buffer and cell death was analyzed by PI incorporation using flow cytometry in 3 independent experiments with 4 replicates per condition. *P < 0.05 comparing between starved PARP-1-knockdown parp-1+/+ MEFs and starved control parp-1+/+ MEFs. (B) Effect of autophagy inhibition with 3-MA on the survival of parp-1+/+ and parp-1−/− MEFs during starvation. 3-MA was added 1.5 h before the HANK buffer as pre-treatment and kept during starvation in both cells lines to maintain the autophagy inhibition. Percentage of survival was obtained by MTT survival assay. Similar survival rates were obtained in three independent experiments with four replicates per condition. (C) Effect of ATG7 knockdown and PARP inhibition on cell death in parp-1+/+ MEFs. Percentage of survival was obtained by MTT assay. Similar survival rates were obtained in three independent experiments with four replicates per condition. While ATG7 silencing prevented cell death, PARP inhibition increased cell death even in cells with limited ability to engage autophagy. (D) Induction of autophagy in MEFs Bax−/−/Bak−/− GFP-LC3 during starvation. Western blot of LC3 conversion (right) and percentage of autophagic cells (left), treatment with rapamycin (100 nM) for 4 h as positive control. DPQ (40 μM) and 3-MA (2 mM) were added 1.5 h before the HANK buffer as pre-treatment and maintained during the starvation. The experiments were repeated three times with similar results. #P < 0.05 comparing between starved Bax−/−/Bak−/− MEFs and 3-MA-treated starved Bax−/−/Bak−/− MEFs or DPQ-treated starved Bax−/−/Bak−/− MEFs. (E) Cell death in MEFs Bax−/−/Bak−/− GFP-LC3 during starvation: levels of cell death measured by PI incorporation using flow cytometry during different times of nutrient deprivation. Three independent experiments with three replicates per condition are represented. (F) Reduction of the percentage of cell death with PARP-1 siRNA (50 nM) during starvation. The right panel shows the silencing of PARP-1 48 h after transfection. Similar results were obtained in 4 experiments with 4 replicates per condition. #P < 0.05, ##P < 0.01 comparing between starved Bax−/−/Bak−/− MEFs and PARP-1-knockdown starved Bax−/−/Bak−/− MEFs. (G) Caspase 3/7 activity in 3T3 MEFs: parp-1+/+ and parp-1−/− 3T3 MEFs cells were starved for 4.5, 6 and 8 h to induce apoptosis. The Caspase - Glo reagent was added directly to cells in 96-well plates and the final volume was 200 μl per well. The assays were incubated at room temperature for 45 min before recording luminiscence in a TECAN infinite 200 Luminometer. Each point represents the average of three wells per condition in three independent experiments. **P < 0.01 comparing between starved parp-1+/+ and parp-1−/− 3T3 MEFs. (H) Activation of caspase 8 in p arp-1−/− MEFs 3T3 under nutrient deprivation. p arp-1+/+ and parp-1−/− MEFs were starved with HANK buffer for 6 h. The fragment of 38 KDa of caspase 8 processed was visualized by western blot and a Jurkat cell total lysate was used as positive control for activation of caspase 8. α-Tubulin was used as loading control. (I) Percentage of apoptotic cells under starvation. p arp-1+/+ and parp-1−/− MEFs were starved for 6 and 8 h and apoptosis was evaluated by double staining of annexin V and PI using flow cytometry. Results are from three independent experiments with two replicates per condition. *P < 0.05 comparing between starved parp-1+/+ and parp-1−/− MEFs.
Autophagy has a pro-survival function after cellular stress such as nutrient withdrawal. This increased cell death following starvation after disabling of PARP-1 could be due to the decreased levels of autophagy. To determine whether autophagy has a cytoprotective role in WT cells, we inhibited autophagy with 3-MA and determined cell viability 8 h after nutrient deprivation (Figure 5B). Blocking autophagy with 3-MA protected parp-1+/+ MEFs against cell death but not PARP-1-deficient cells, suggesting that WT cells died mainly as consequence of autophagy after prolonged starvation, while the inactivation of PARP-1 may lead to non-autophagic cell death. Silencing of the autophagy gene ATG7 strongly delayed cell death in MCF-7 cells subjected to nutrient starvation while the treatment with the PARP inhibitor PJ34 still increased cell death of ATG7-silenced/autophagy-deficient cells, suggesting that autophagy-derived cell death is not involved in the accelerated cell demise caused by PARP inhibition (Figure 5C).
To analyze the type of cell death that takes place in the absence or after inhibition of PARP-1 during starvation, we used a double-knockout cell line for Bax and Bak. Bax and Bak are two essential proteins in the apoptotic process and these cells cannot undergo mitochondrial outer membrane permeabilization (MOMP) that usually occurs during apoptosis 22. Additionally, this cell line is stably transfected with GFP-LC3. To determine the levels of autophagy, we starved these cells for different times up to 120 min (Figure 5D). Bax/Bak double-mutant cells displayed LC3 processing during starvation (Figure 5D) and the percentages of cells with the typical GFP-LC3 pattern (Figure 5D) were similar to parp-1+/+ MEFs. Moreover, co-treatment with 3-MA or the PARP inhibitor DPQ reduced the percentage of autophagic cells, as also observed for parp-1+/+ MEFs (Figures 2A and 5D). To determine to what extent the apoptotic component contributes to the increased cell death in the absence of PARP-1, we silenced PARP-1 in Bax−/−/Bak−/− GFP-LC3 MEFs by siRNA. Knockdown of PARP-1 in the context of bax/bak knockout decreased the levels of cell death during nutrient starvation (Figure 5E and 5F), suggesting that the gain in cell death after inactivation of PARP-1 (in the Bax/Bak WT context (Figure 5A-5C)) has an apoptotic component. To confirm this result, we have measured caspase 3/7 and caspase-8 activation by fluorimetric assay and western blot, respectively. Time course of caspase 3/7 activation was significantly increased in parp-1−/− cells (Figure 5G); and caspase-8 processing did not take place in parp-1+/+ MEFs (Figure 5H). Moreover, by quantitation of pycnotic nuclei (data not shown) as well as double staining with annexin V and PI (Figure 5I), apoptotic cells were determined to be significantly increased in the absence of PARP-1. These data confirm that parp-1−/− MEFs with delayed autophagy enter into apoptosis after several hours of starvation. Thus, upon prolonged starvation PARP-1 WT cells die by autophagy-dependent cell death while PARP-1 knockout cells die mainly by apoptosis.
Impaired starvation-induced autophagy in PARP-1-deficient mice
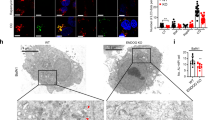
In order to analyze the in vivo consequences of PARP-1 ablation on autophagy, we starved pups from both parp-1+/+ and parp-1−/− mice for 4 h. TEM (transmission electron microscopy) analysis showed that liver from fed PARP-1-proficient mice displayed characteristic abundant, well-structured mitochondria as well as lipid droplets. After starvation, autophagy-derived ultrastructural changes include concentrical membrane structures engulfed in autophagosomes, ER dilation and also accumulation of lipid droplets, which were already found in non-starved liver (Figure 6A, upper panels). Concentrical membrane structures reflect degradation of membranous cellular components that rearrange in membranous whorls called myelin figures. By contrast, TEM images of parp-1−/− liver cells showed remarkable differences in ultrastructural morphology in both fed and starved pups with a well-organized ER and the absence of concentrical membranes structures and lipid droplets (Figure 6A, lower panels). To further support this observation, we measured changes in LC3-I levels (no LC lipidation was detected in this experiment) in liver samples from parp-1+/+ and parp-1−/− pups subjected or not to starvation. Reduction in LC3-I was much more pronounced in livers from parp-1+/+ mice (Figure 6B).
Deficient liver autophagy in starved PARP-1 knockout mice. 2 h after birth pups were separated from their mother and starved for 4 h according to procedures approved by the bioethics committee of the CSIC. Pups were sacrificed and livers were removed and fixed for TEM (A) or lysed for western blot analysis (B). A, Autophagosomes showing concentric membrane structures; ER, endoplasmic reticulum; L, lipid droplets; Ly, lysosomes; N, nucleus. (B) Immunoblot analysis of endogenous LC3 conversion in control and starved parp-1+/+ and parp-1−/− neonatal mice. β-actin was used as loading control. Similar results were obtained in 3 independent experiments. (C) Lipids droplets in starved parp-1+/+ and parp-1−/− MEFs. Cells were starved for 1 or 2 h and numbers of BODIPY-positive vesicles per cell are shown. Treatment with 20 μM CQ for 2 h was used as positive control. *P < 0.05 comparing between parp-1+/+ MEFs starved for 1 h and those starved for 2 h.
The intracellular storage and utilization of lipids are critical to maintain cellular energy homeostasis. Cellular lipids are stored as triglycerides in lipid droplets in the fed liver and hydrolyzed into fatty acids for energy production, which is also one of the initial responses to starvation. A second cellular response to starvation is the induction of autophagy, which delivers intracellular proteins and organelles sequestered in double-membrane vesicles to lysosomes for degradation and use as an energy source. To further confirm the finding that parp-1−/− liver cells did not accumulate lipid droplets in response to starvation (Figure 6A and 6B), we used the lipid fluorescence dye BODIPY® to label lipid droplets and showed that BODIPY-positive vesicles were strongly reduced in parp-1−/− cells (Figure 6C). Thus, in the absence of PARP-1, liver from neonates has impaired autophagic response as has been reported in knockout mice for genes involved in the core complex of autophagy 23, 24.
Discussion
Genetic and environmental factors modulate the response of multicellular organisms to stress and the maintenance of tissue homeostasis and highly integrated response patterns are found in many organisms, but the means by which so many diverse pathways, critical for cellular, tissue and ultimately for organism survival, are coordinated, has yet to be elucidated. In this study, we show that optimal induction of autophagy induced by nutrient deprivation requires PARP-1 activation. Our results clearly show the lack of AMPK activation after disabling of PARP-1. These findings are in agreement with previous results showing an interaction between PARP-1 and AMP metabolism related to the hydrolysis of ADP-ribose 25. In the absence of PARP-1 or after its inhibition/silencing, ATP levels are not reduced as much as in WT cells (Figure 4A); consequently AMPK remains inactive and does not signal for mTOR inactivation, leading to impaired autophagy. At present the link between PARP-1 activation and AMPK has not been established. One speculative possibility is that ROS-induced DNA damage and PARP-1 overactivation caused mitochondrial dysfunction and Ca2+ release, thereby activating AMPK 14. Other possibility that may explain how PARP-1 connects with the AMPK/mTOR signaling (besides the maintenance of the energy status) is that PARP inactivation leads to unefficient ATM response that has been reported to be involved in the activation of AMPK 26, 27.
Our data suggest that the PARP-1 is an important in vivo regulator of autophagy and provide a link between PARP-1 function and the overall cellular response to nutrient shortage. The results demonstrate that nutrient starvation, ROS production and DNA damage lead to PARP-1 activation, which is needed for cells to engage starvation-induced autophagy.
We also show that the role of PARP-1 in starvation-induced autophagy is related to its ability to sense DNA damage and deplete energy stores after its overactivation, but we cannot exclude the possibility of perturbation in Ca2+ flux after PARP-1 ablation upstream of the mitochondria leading to altered ATP synthesis and AMPK activation 28. Moreover, we have also analyzed the in vivo consequences of PARP-1 inactivation in starvation-induced autophagy. Starvation induces hepatic autophagy and increases the delivery of fatty acids to the liver from lipolysis of adipose tissue. Electron microscopy revealed that starvation increased the frequency of lipid droplets with increased density and asymmetrically localized multi-membrane structures. The crucial role of neonatal autophagy was clearly demonstrated by targeted inactivation of the autophagy-related genes ATG5 and ATG7 23, 29. Mice deficient in these genes were apparently normal at birth, except for a slightly lower body weight than control (approximately 10% in ATG5-null and 18% in ATG7-null mice), but died within 1 day after birth. One of the phenotypes of PARP-1 knockout mice was that the average litter size was smaller (about 40%) than that of parp-1+/+ mice 30. It would be interesting to further explore whether the reduced litter size of PARP-1 knockout mice is related to the changes in autophagy.
ROS have been reported to be a hallmark of autophagy in a number of cell types and experimental settings, including nutrient starvation 9. Although autophagy after nutrient deprivation has a pro-survival function, our findings support that PARP-1 is necessary for cells to persist in autophagy following starvation when nutrient deprivation is maintained for a long time (Figure 5B). An interesting finding in our study is that suppression of PARP activation by different means leads to impaired autophagy and eventually to increased cell death. Indeed, exposure of PARP-1-deficient cells to a longer starvation period (6 and 8 h) resulted in increased cell death (data not shown). Additionally we have defined the mechanism by which PARP-1 suppression accelerates cell death using the apoptosis-deficient cell line with double knockout for bax and bak. We found that starvation leads Bax−/−/bak−/− cells to autophagy and cell death, suggesting that apoptosis is not the main pathway of cell demise triggered by starvation. Nonetheless, suppression of PARP-1 in this Bax−/−/bak−/− context substantially decreased cell death, contrary to what we observed in apoptosis-proficient cells. One major observation in this study is that a physiological trigger of autophagy, such as nutrient deprivation, is able to induce DNA damage through the generation of ROS. Genotoxic stress has been reported to repress mTOR in response to oxidative stress caused by ROS through a cytoplasmic signaling node for LKB1/AMPK/TSC2 activation in response to oxidative stress 31. The COMET assay and histone γ-H2AX accumulation confirmed the persistence of damaged DNA and the level of initial damage corresponded with the cell's ability to initiate autophagy. Treatment with antioxidant NAC prevented DNA damage and mTOR inactivation, and slowed down autophagy. Why are PARP-1-deficient cells prone to die by apoptosis following nutrient deprivation? One interesting possibility is suggested by the results in Figure 3C-3E: PARP-1 mutant cells display a defective DNA repair during starvation; thus, the cells choose to undergo apoptosis to avoid the harm of bearing unrepaired DNA; in a tumor context, where oxygen and nutrients are limited, this delayed autophagy (together with the tendency to die by apoptosis in the absence of PARP-1), might have benefits by preventing necrosis, ROS and inflammatory burst from tumor growth.
The ultimate reason why PARP-1 deficiency or inactivation leads to inhibition of ROS generation is not completely understood. Interestingly, early reports have also shown that alterations in mitochondrial function during oxidant-mediated cytotoxicity, are related to PARP-1 activation rather than to direct effects of the oxidants on the mitochondria 12. ER and mitochondrial Ca2+ signaling is a key mediator of cell's bioenergetic regulation and cell death. Constitutive InsP(3)R Ca2+ signaling is required for autophagy suppression in cells under non-starvation conditions. It has been shown that after nutrient deprivation cells become metabolically compromised due to diminished mitochondrial Ca2+ uptake 28. Very recently PARP-1 and PARG (poly(ADP-ribose)glycohydrolase) have been reported to regulate Ca2+ influx through TRPM2 32 and a reduction in Ca2+ was observed after abrogation of PARP-1. In this study by Blenn et al. 32, they reported that the increased Ca2+ flux following H2O2 treatment leads to caspase activation and cleavage of mitochondrial AIF, which then translocates to the nucleus to cause DNA fragmentation, chromatin condensation and cell death. Clearly, the level of ROS-derived cyotoxicity and PARP-1 activation differs in both settings: while H2O2 treatment produces an overwhelming burst in oxidant mediators, starvation leads to a mild ROS generation allowing the cell not to undergo direct cell death but to engage the pro-survival autophagy. In support of that, Scherz-Shouval et al. 9 demonstrated that ROS in starvation-induced autophagy has a pro-survival function. They showed that oxidative conditions are essential for autophagy and that the increase in ROS is both local and reversible during starvation, which is not deleterious to cells and serves to oxidize a specific target. By contrast, massive ROS production during oxidative stress will lead to ROS-derived cell death by autophagy 33. Once starvation has initiated the commitment to autophagy, cells abrogated of PARP-1 undergo a “slow” autophagy that is eventually resolved by increased cell death, particularly (but not exclusively) apoptosis. It has been previously shown that PARP inhibition can shift the necrotic cell death to apoptosis after exposure to oxidative stress 34. In agreement with that, our results show that the cell death observed in the absence of PARP-1 after starvation (leading to oxidative stress) is due to increased apoptosis. In this context PARP-1 is needed for cells to undergo pro-survival autophagy.
Our model is presented in Figure 7 and could be summarized as follows: after nutrient deprivation mitochondrial metabolism is rapidly shifted, leading to ROS production and ATP drop. An elevation in the AMP/ATP ratio activates the nutritional sensor kinase AMPK, whose activation leads to mTORc1 inhibition, allowing the commitment to autophagy. In parallel, ROS production induces DNA damage and PARP-1 overactivation, contributing to the feedback loop to decrease ATP through the consumption of NAD+. In this scenario, the axis ROS/AMPK/mTOR and ROS/DNA damage/PARP-1 activation synergize to optimize the cell's response to nutrient deprivation by inducing pro-survival autophagy (Figure 7A). In the absence of PARP-1, ROS production, energy drop and AMPK activation are diminished and shut-off of the feedback loop responsible for massive energy depletion eventually slows down autophagy. Alternatively, the cells die through apoptosis due to suboptimal autophagy commitment (Figure 7B).
(A) PARP-1 is a positive modulator of starvation-induced autophagy. After nutrient starvation cells activate autophagy through the activation of AMPK/inhibition of mTOR. Upstream events involve energy depletion, ROS production and DNA damage. Under this condition, PARP-1 overactivation leads to ATP depletion, acting as a feedback loop to reactivate autophagy. This stress signal when maintained eventually leads to cell death through autophagy. (B) PARP inactivation delays autophagy and favors apoptosis. In the absence of PARP-1 or after PARP inactivation, ROS levels decreases and ATP drop is reduced. As a consequence, the feedback loop reactivated by PARP-1 does not take place, and apoptosis is triggered as a mode of cell death.
Autophagy is a potent tumor suppressive mechanism, presumably due to its essential contribution to the maintenance of genomic stability 35, the avoidance of excessive ROS generation 36 and its participation in cellular senescence 37, which constitutes a barrier against oncogenesis. Accordingly, multiple genes that are required for the induction/execution of autophagy are potent tumor suppressors, including PTEN, TSC1, TSC2, LKB1, ATG4, Beclin-1, UVRAG, and BH3-only proteins of the Bcl-2 family 38. Here, we revealed the importance of PARP for the autophagic process in a physiologic setting following nutrient starvation. PARP-1 inhibitors are entering clinical trials for different types of cancer. Whether the ability of PARP inhibitors to favor apoptotic cell death during cellular stress, such as shortage of nutrients (which very often the case in tumor microenvironnement), could also be exploited in antitumor therapy by its contribution to autophagy, remains an intriguing possibility for further investigation.
Materials and Methods
Cell culture and treatment
Immortalized MEFs 3T3, derived from both WT and PARP-1 KO mice, Bax−/−/Bak−/− GFP-LC3 MEFs 3T3, g361 and HT144 cell lines were cultured in Dulbecco's modified Eagle's medium supplemented with 10% inactive fetal bovine serum (FBSi, Gibco Invitrogen) at 37 °C in a humidified 5% CO2 atmosphere. MCF7-GFP-LC3 cells were cultured in RPMI-1640 GlutaMAX (GIBCO Invitrogen) with 6% FBSi. Cells were starved with balanced HANK buffer without amino acids (NaCl 140 mM, KCl 5 mM, MgCl2-6H2O 1.3 mM, CaCl2-2H2O 2 mM, HEPES 10 mM, D-glucose 5 mM) for different time periods.
For western blot, cells were plated in six-well plates with a density of 4 × 105 cells per plate and treated with HANK buffer the next day. For the assessment of cell death, cells were plated in 24-well plates with a density of 3.5 × 104 cells per well and in six-well plates with 2.5 × 105 cells per well. To count the number of vesicles per cell, cells were plated in six-well with cell density of 4 × 104 cells per well on coverslips and starved in the next day for different time periods. Fluorescence microscopy analysis was performed with a Zeiss microscope.
The autophagy inhibitor 3-MA (M9281, Sigma-Aldrich, St Louis, MO, USA) was dissolved in culture medium and stored at −20 °C (10 mM). PARP-1 inhibitor DPQ (3,4-Dihydro-5-[4-(1-piperidinyl)butoxy]-1(2H)-isoquinolinone]) and PJ34 were from Alexis Biochemicals. DPQ was dissolved in DMSO and stored at −20 °C. Cells were pre-treated with 40 μM DPQ or 10 μM PJ34 for 1.5 h before starvation and maintained all the time during the experiment.
Cell viability assay
The levels of cell death in parp-1+/+ and parp-1−/− MEFs were determined using trypan blue exclusion assay (93595, Fluka, St Louis, MO, USA) (a), MTT (b) and PI (c). (a) Trypan blue: 3T3 cells were seeded at 3.5 × 104 cells per well in 24-well plates and incubated overnight at 37 °C. After starvation with HANK buffer, cells were washed and trypsinized. The cellular pellet was dissolved in 50 μl of trypan blue solution and the number of viable cells was counted under a normal microscope. (b) MTT assay (3-(4, 5-Dimethylthiazol-2-yl)-2,5-diphenyl Tetrazolium Bromide) was performed using Cell Proliferation Kit I (MTT, 1-65-007, Roche, Germany) following the manufacturer's instructions. (c) PI was used as exclusion staining and a FACScalibur flow cytometer with the Cell Quest software (BD Biosciences) was used to perform analysis.
Immunofluorescence
Immunostaining for PAR was performed on cells grown on glass coverslips and fixed in ice-cold methanol-acetone (1:1) for 10 min. PAR was detected by immunofluorescence, using monoclonal antibody (Trevigen) and FITC-conjugated goat anti-mouse immunoglobulin (Sigma-Aldrich). Foci of γ-H2AX were detected with a monoclonal antibody for H2AX histone (Upstate, #05-636, clone JBW103) and FITC-conjugated goat anti-mouse antibody. Nuclear counterstaining with DAPI was performed after removal of excess secondary antibody. Immunostaining was visualized with a Zeiss fluorescence microscope.
Quantification of lipid droplets by fluorescence microscopy
parp-1+/+ and parp-1−/− MEFs were seeded (4 × 104 cells per well) in six-well plates on glass coverslips. Cells were starved for 1 and 2 h and fixed with paraformaldehyde solution (4%, wt/vol in 1× PBS with 2% sucrose) for 10 min at room temperature. Lipid droplets were labeled with BODIPY® 493/503 (Invitrogen) for 5 min at room temperature and visualized under a Zeiss fluorescence microscope. Nuclear counterstaining with DAPI was performed after probing. Incubation with 20 μM of chloroquine for 2 h was used as positive control of accumulation of lipids droplets.
Western blot analysis
After the nutrient starvation with HANK buffer, cells were washed twice with PBS and resuspended in 70 μl of lysis buffer (50 mM Tris-HCl pH 8, 0.1 mM EDTA, 0.5% Triton X-100, 12.5 mM β-Mercaptoethanol) for 45 min on ice. Pellet was eliminated and the supernatant was stored at −20 °C. Proteins were resolved on 10% SDS-polyacrylamide gels and transferred onto PVDF membrane (Bio-Rad). The blot was blocked with 5% milk powder in 1× PBS and 0.1% Tween-20 for 60 min, washed with PBS/Tween, and incubated overnight with the different antibodies, anti-poly(ADP-ribose) (TREVIGEN, 20591E10, My, USA), anti-PARP-1(C2-10 mouse, ALEXIS, LA), anti-LC3 (NanoTools, clone 5F10, Ref 03231-100/LC3-5F10), anti-Atg7 (Cell Signaling Technology, Beverly, MA, USA), anti-phospho-p70S6K (Cell Signaling Technology), anti-p70S6K (Cell Signaling Technology), anti-phospho-AMPKα (Thr172) (Cell Signaling Technology), anti-AMPKα (Cell Signaling Technology) and anti-caspase 8 (BD Pharmingen). α-Tubulin (Sigma-Aldrich), β-actin (Sigma-Aldrich) and GAPDH (Santa Cruz Biotechnology) were used as loading controls. Bands were visualized by ECL-Plus (Amersham Biosciences) and the pictures were taken with ChemiDoc XRS imaging system (Bio-Rad) or medical X-ray films (AGFA).
ATP determination
Intracellular ATP was measured using a luciferin/luciferase-based assay (ATP Bioluminescent Assay Kit, Sigma-Aldrich) following the manufacturer's guidelines. A standard curve was generated with known concentrations of ATP and used to calculate sample ATP concentrations. Protein concentration was determined using Bradford protein assay reagents (Bio-Rad). The content of ATP was normalized for protein content.
Autophagy assay
GFP-LC3-expressing cells have been used to demonstrate the induction of autophagy. The GFP-LC3 expression vector was kindly supplied by Dr T Yoshimori (National Institute for Basic Biology, Okazaki, Japan). parp-1+/+ and parp-1−/− MEFs were transiently transfected with this vector together with jetPEITM (Polyplus transfection, Illkirch, France) according to the manufacturer's protocol. The assay was performed on cells grown on glass coverslips (4 × 104 cells per well) in six-well plates and after the different treatment with HANK buffer, cells were washed three times with sterile PBS and fixed with paraformaldehyde solution (4%, wt/vol in 1× PBS with 2% sucrose) for 10 min at room temperature. To determine LC3 localization, GFP-LC3-transfected cells were observed under a Zeiss fluorescence microscope. To determine LC3-II translocation in parp-1+/+ and parp-1−/− MEFs, we performed western blot of LC3-I and its proteolytic derivative LC3-II (18 and 16 kDa, respectively) using a monoclonal antibody against LC3 (NanoTools, clone 5F10, Ref 03231-100/LC3-5F10).
Rapamycin (553210 Calbiochem, Germany) and Concanamycin A (C9705, Sigma-Aldrich) were used as autophagy inducers. Rapamycin was used at 100 nM for 4 h and Concanamycin A at 4 nM for 4 h.
Apoptosis assay
In addition to caspase 8 cleavage, apoptosis was determined by two different methods: (1) Pyknotic nuclei: Cells were fixed by paraformaldehyde (4%, wt/vol in 1× PBS with 2% sucrose) for 10 min at room temperature and the number of cells with nuclear apoptotic morphology was determined by DAPI staining after 6 and 8 h of starvation, using a Zeiss fluorescence microscope. Treatment with 0.5 μg/ml cycloheximide (CHX C4859, Sigma-Aldrich) for 8 h was used as positive control of nuclear apoptotic morphology. (2) Annexin V/IP: parp-1+/+ and parp-1−/− MEFs (5 × 105 cells per well) were cultured in 6-well plates and starved for 6 h. After starvation, cells were washed twice with PBS, trypsinized and centrifuged at 1 600 rpm for 5 min. The pellets were re-suspended in AnnexinV/IP solution (Roche Applied, Germany) according to the manufacturer's instruction and maintained in the dark at 37 °C for 15 min. Apoptotic cells were evaluated in a FACScalibur flow cytometer with the Cell Quest software (BD Biosciences, NJ, USA). Staurosporin (6942, Sigma-Aldrich; 3 μM) for 8 h was used as a positive control of apoptosis induction.
Caspase 3/7 activity in 3T3 MEFs: parp-1+/+ and parp-1−/− MEFs were starved for 4.5, 6 and 8 h to induce apoptosis. The Caspase-Glo reagent was added directly to cells in 96-well plates; the final volume was 200 μl per well. The cells were incubated at room temperature for 45 min before recording luminiscence in a TECAN infinite 200 Luminometer. Each point represents the average of three wells per condition in three independent experiments. The “no-cell” blank control value has been substracted from each point. STS (2 μM for 3 h) was used as positive control of caspase activation. Data generated in apoptotic cells with STS are not shown. **P < 0.01 comparing between starved parp-1+/+ and parp-1−/− MEFs.
RNA interference
Cells were transfected with the indicated siRNAs (50 nM) using Dharmafect transfection agent (Dharmacon Research, CO, USA) according to the manufacturer's guide. siRNAs corresponding to the cDNA sequences were: ATG7 no.1 from Invitrogen, human PARP-1 from Ambion Applied Biosystems and mouse PARP-1 from Santa Cruz Biotechnology.
48 h after transfection, cells were treated as described and observed under a Zeiss fluorescence microscope or the proteins were extracted.
ROS and DNA damage determination
ROS production was measured by flow cytometry in an Epics Elite ESP cytometer (Coulter) using DCFDA (35845, FLUKA; 8 mg/ml in DMSO), a specific probe to ROS. 3T3 cells were seeded at 3.5 × 105 cells per well in 6-well plates and incubated overnight at 37 °C. Cells were incubated with the probe for 30 min before the end of the deprivation and washed twice with PBS, trypsinized and the ROS production was analyzed by flow cytometry.
DNA damage was analyzed using two different methods: (1) COMET assay: DNA damage was quantified using COMET Assay kit (R&D Systems, Trevigen, MD, USA) with some modifications. 1 × 105 cells/ml were mixed with molten LM agarose at 37 °C at a ratio of 1:10 (vol/vol) and pipetted onto a COMET slide. The slides were placed for 10 min in the dark at 4 °C and were immersed in pre-chilled lysis solution. The slides were then removed from lysis buffer, washed in TBE buffer and transferred to a horizontal electrophoresis chamber. Voltage (1 V/cm) was applied for 20 min. After washed in distilled water, the slides were immersed in 70% ethanol for 5 min and allowed to air dry. Slides were stained with SYBR Green and then analyzed by fluorescence microscopy. 70-90 cells were evaluated in each sample using the COMET Assay Software Project (CASP software). DNA damage was quantified by measuring the TM calculated as percentage of DNA in the tail × tail length. (2) phospho-γ-H2AX: To visualize foci of γ-H2AX, we performed immunofluorescence with a monoclonal antibody for H2AX histone (Upstate, #05-636, clone JBW103) and FITC-conjugated goat anti-mouse antibody. Nuclear counterstaining with DAPI was performed after removal of excess secondary antibody. Immunostaining was visualized with a Zeiss fluorescence microscope. Western blot analysis of γ-H2AX phosphorylation was performed with the same antibody. Bands were visualized by ECL-Plus (Amersham Biosciences) and the pictures were taken with ChemiDoc XRS imaging System (Bio-Rad) or medical X-ray films (AGFA).
Electron microscopy
Animal experimental protocols were reviewed and approved by the Ethical Committee of the Spanish Council of Scientific Research (CSIC). PARP-1 WT and knockout 30 neonatal mice were used to determine the differences in autophagy induction during starvation in hepatic tissue. Neonatal mice were separated from the mother in the first 4 h after birth. Livers of WT and PARP-1 knockout mice were extracted and washed with PBS, prefixed for 30 min in a fixation solution (0.1 M cacodilate buffer pH 7.4 and osmium tetraoxyde) for 60 min at 4 °C. After this treatment, tissues were washed with MilliQ water and the samples were stained with uranil acetate. The ultrathin sections were cut with a diamond knife in an ultramicrotome (Reichert Ultracut S). The samples were analyzed in a TEM Zeiss 902 with 80 KV of voltage acceleration (CIC-UGR).
References
Levine B, Kroemer G . Autophagy in the pathogenesis of disease. Cell 2008; 132:27–42.
Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000; 19:5720–5728.
Schreiber V, Dantzer F, Ame JC, de Murcia G . Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 2006; 7:517–528.
David KK, Andrabi SA, Dawson TM, Dawson VL . Parthanatos, a messenger of death. Front Biosci 2009; 14:1116–1128.
Wang Y, Dawson VL, Dawson TM . Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos. Exp Neurol 2009; 218:193–202.
Munoz-Gamez JA, Rodriguez-Vargas JM, Quiles-Perez R, et al. PARP-1 is involved in autophagy induced by DNA damage. Autophagy 2009; 5:61–74.
Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ . The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem 1997; 243:240–246.
Krishnakumar R, Kraus WL . The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell 2010; 39:8–24.
Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z . Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 2007; 26:1749–1760.
Chen Y, Azad MB, Gibson SB . Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ 2009; 16:1040–1052.
LeBel CP, Ischiropoulos H, Bondy SC . Evaluation of the probe 2′,7′-dichlorofluorescin as an indicator of reactive oxygen species formation and oxidative stress. Chem Res Toxicol 1992; 5:227–231.
Virag L, Salzman AL, Szabo C . Poly(ADP-ribose) synthetase activation mediates mitochondrial injury during oxidant-induced cell death. J Immunol 1998; 161:3753–3759.
Hay N, Sonenberg N . Upstream and downstream of mTOR. Genes Dev 2004; 18:1926–1945.
Hoyer-Hansen M, Jaattela M . AMP-activated protein kinase: a universal regulator of autophagy? Autophagy 2007; 3:381–383.
Shaw RJ, Bardeesy N, Manning BD, et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 2004; 6:91–99.
Gogvadze V, Orrenius S, Zhivotovsky B . Mitochondria in cancer cells: what is so special about them? Trends Cell Biol 2008; 18:165–173.
Tomitsuka E, Kita K, Esumi H . The NADH-fumarate reductase system, a novel mitochondrial energy metabolism, is a new target for anticancer therapy in tumor microenvironments. Ann N Y Acad Sci 2010; 1201:44–49.
Kroemer G, Levine B . Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol 2008; 9:1004–1010.
Levine B, Yuan J . Autophagy in cell death: an innocent convict? J Clin Investig 2005; 115:2679–2688.
Baehrecke EH . Autophagy: dual roles in life and death? Nat Rev 2005; 6:505–510.
Boya P, Gonzalez-Polo RA, Casares N, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol 2005; 25:1025–1040.
Caro-Maldonado A, Tait SW, Ramirez-Peinado S, et al. Glucose deprivation induces an atypical form of apoptosis mediated by caspase-8 in Bax-, Bak-deficient cells. Cell Death Differ 2010; 17:1335–1344.
Komatsu M, Waguri S, Ueno T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. The J Cell Biol 2005; 169:425–434.
Lee IH, Cao L, Mostoslavsky R, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA 2008; 105:3374–3379.
Formentini L, Macchiarulo A, Cipriani G, et al. Poly(ADP-ribose) catabolism triggers AMP-dependent mitochondrial energy failure. J Biol Chem 2009; 284:17668–17676.
Aguilar-Quesada R, Munoz-Gamez JA, Martin-Oliva D, et al. Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition. BMC Mol Biol 2007; 8:29.
Haince JF, Kozlov S, Dawson VL, et al. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J Biol Chem 2007; 282:16441–16453.
Cardenas C, Miller RA, Smith I, et al. Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell 2010; 142:270–283.
Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature 2004; 432:1032–1036.
de Murcia JM, Niedergang C, Trucco C, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci USA 1997; 94:7303–7307.
Nakada D, Saunders TL, Morrison SJ . Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature 2010; 468:653–658.
Blenn C, Wyrsch P, Bader J, Bollhalder M, Althaus FR . Poly(ADP-ribose)glycohydrolase is an upstream regulator of Ca2+ fluxes in oxidative cell death. Cell Mol Life Sci 2011; 68:1455–1466.
Yu L, Alva A, Su H, et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science 2004; 304:1500–1502.
Virag L, Scott GS, Cuzzocrea S, Marmer D, Salzman AL, Szabo C . Peroxynitrite-induced thymocyte apoptosis: the role of caspases and poly (ADP-ribose) synthetase (PARS) activation. Immunology 1998; 94:345–355.
Karantza-Wadsworth V, Patel S, Kravchuk O, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev 2007; 21:1621–1635.
Mathew R, Karp CM, Beaudoin B, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009; 137:1062–1075.
Young AR, Narita M, Ferreira M, et al. Autophagy mediates the mitotic senescence transition. Genes Dev 2009; 23:798–803.
Maiuri MC, Criollo A, Kroemer G . Crosstalk between apoptosis and autophagy within the Beclin 1 interactome. EMBO J 2010; 29:515–516.
Acknowledgements
We acknowledge Laura López for her technical assistance. JMRV is recipient of a predoctoral fellowship financed by the program JAE-Pre of CSIC and was also funded by CSIC with a Short-Term Fellowship to stay at the Danish Cancer Society Institute of Cancer Biology in Copenhagen, Denmark. This work was supported by Ministerio de Ciencia e Innovación (SAF2006-01094 and SAF2009-13281-C02-01), Fundación La Caixa (BM06-219-0) and Junta de Andalucía (P07-CTS-0239) to FJO; RTICC (RD06/0020/0068) to ALR.
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
parp-1+/+ MEFs 3T3 and parp-1−/− transfected transiently with GFP-LC3. (PDF 853 kb)
Supplementary information, Figure S2
Effect of the chemical inhibition (3MA) and of ATG7 silencing on autophagy during starvation in parp-1+/+ and parp-1+/+ MEFs 3T3 (PDF 797 kb)
Supplementary information, Figure S3
(A) Lysosome fusion with autophagosomes is not affected in parp-1−/− cells. (PDF 448 kb)
Supplementary information, Figure S4
Reconstituted expression of PARP-1 in parp-1−/− MEFs 3T3 restored the autophagy levels. (PDF 1009 kb)
Supplementary information, Figure S5
PAR polymer production confirms PARP-1 over-activation during starvation. (PDF 913 kb)
Supplementary information, Figure S6
γ - H2AX Staining during starvation in parp-1+/+ MEFs 3T3 and parp-1-A. (PDF 1067 kb)
Supplementary information, Figure S7
MCF7 cells stably transfected with GFP-LC3 were starved in the absence or presence of 10 nM 3MA. (PDF 500 kb)
Supplementary information, Figure S8
Delay of autophagy in a deficient DNA repair model (PDF 803 kb)
Rights and permissions
About this article
Cite this article
Rodríguez-Vargas, J., Ruiz-Magaña, M., Ruiz-Ruiz, C. et al. ROS-induced DNA damage and PARP-1 are required for optimal induction of starvation-induced autophagy. Cell Res 22, 1181–1198 (2012). https://doi.org/10.1038/cr.2012.70
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2012.70
Keywords
This article is cited by
-
Cannabidiol-induced crosstalk of apoptosis and macroautophagy in colorectal cancer cells involves p53 and Hsp70
Cell Death Discovery (2023)
-
A “three musketeers” tactic for inclining interferon-γ as a comrade-in-arm to reinforce the synergistic-tumoricidal therapy
Nano Research (2022)
-
Mitochondrial changes in fish cells in vitro in response to serum deprivation
Fish Physiology and Biochemistry (2022)
-
Endonuclease G promotes autophagy by suppressing mTOR signaling and activating the DNA damage response
Nature Communications (2021)
-
NRF2-GPX4/SOD2 axis imparts resistance to EGFR-tyrosine kinase inhibitors in non-small-cell lung cancer cells
Acta Pharmacologica Sinica (2021)