Abstract
The MLL-AF4 fusion gene is a hallmark genomic aberration in high-risk acute lymphoblastic leukemia in infants. Although it is well established that MLL-AF4 arises prenatally during human development, its effects on hematopoietic development in utero remain unexplored. We have created a human-specific cellular system to study early hemato-endothelial development in MLL-AF4-expressing human embryonic stem cells (hESCs). Functional studies, clonal analysis and gene expression profiling reveal that expression of MLL-AF4 in hESCs has a phenotypic, functional and gene expression impact. MLL-AF4 acts as a global transcriptional activator and a positive regulator of homeobox gene expression in hESCs. Functionally, MLL-AF4 enhances the specification of hemogenic precursors from hESCs but strongly impairs further hematopoietic commitment in favor of an endothelial cell fate. MLL-AF4 hESCs are transcriptionally primed to differentiate towards hemogenic precursors prone to endothelial maturation, as reflected by the marked upregulation of master genes associated to vascular-endothelial functions and early hematopoiesis. Furthermore, we report that MLL-AF4 expression is not sufficient to transform hESC-derived hematopoietic cells. This work illustrates how hESCs may provide unique insights into human development and further our understanding of how leukemic fusion genes, known to arise prenatally, regulate human embryonic hematopoietic specification.
Similar content being viewed by others
Introduction
The mixed-lineage leukemia (MLL) gene fuses to generate chimeric genes with over 70 partners in human leukemia 1. Infant pro-B acute lymphoblastic leukemia (ALL) harboring the fusion MLL-AF4 is characterized by a very brief latency and dismal prognosis, raising the question of how this infant cancer evolves so quickly 2, 3. Moreover, the exceptionally high concordance rate of this leukemia in monozygotic twin infants, approaching 100% 4, suggests that all the necessary genetic events required for leukemogenesis are accomplished prenatally 5.
MLL-AF4-induced leukemogenesis has been particularly difficult to model and bona fide MLL-AF4 disease human models do not exist to date. Our understanding of transformation by MLL fusions and their mode of action comes from murine models in which leukemias do not recapitulate the human disease faithfully 6, 7, 8. These findings raise important questions about MLL-AF4+ leukemia and suggest that these mouse models may be missing some essential ingredients of leukemogenesis during early human development. It could be argued that the lack of a bona fide MLL-AF4 disease model may be due to: (i) a cell in a wrong developmental stage was targeted in the murine approaches; (ii) the impact of other secondary hits has not been properly addressed; or (iii) MLL-AF4 exerts its transforming function preferentially in human cells, indicating that questions regarding the MLL-AF4 pathogenesis have to be addressed using ontogenically primitive human stem cells. Among these, postnatal (cord blood (CB)-derived) CD34+ hematopoietic stem/progenitor cells (HSPCs) or prenatal (fetal- or embryonic-derived) cells represent potential ontogenically early target cells in MLL-AF4 pathogenesis.
Very recently, Montes et al. 9 explored for the first time the in vitro and in vivo developmental impact of MLL-AF4 on the fate of human neonatal CD34+ HSPCs. The expression of MLL-AF4 in human CB-derived HSPCs augmented the in vivo multilineage hematopoietic engraftment and homing, the in vitro clonogenic potential and enhanced their proliferation. However, MLL-AF4 was not sufficient for leukemogenesis on its own, indicating that additional hits are required to develop leukemia or that CB-HSPCs do not constitute the appropriate target for MLL-AF4-mediated ALL.
Human embryonic stem cells (hESC) are envisioned to become a powerful tool for modeling different aspects of human diseases that cannot otherwise be addressed by patient sample analyses or mouse models 10, 11. The fact that leukemogenesis manifests as altered cell differentiation suggests that hematopoietic-directed differentiation of hESCs could become a promising human-specific strategy to study the onset of hematopoiesis, particularly the emergence of the earliest events leading to the specification of both normal and abnormal hematopoietic tissue 12. During hESC differentiation, a population of primitive hemogenic precursors arises that is uniquely responsible for hematopoietic and endothelial development 13, 14, 15. Interestingly, MLL fusions have also been implicated in endothelial cell maturation 16 and endothelial dysfunction has recently been linked to disease outcome in childhood leukemias 17. We have thus explored the developmental impact of MLL-AF4 on the fate of hESCs and hESC-derived hemogenic precursors. We posed the following questions. First, what is the developmental impact of MLL-AF4 on the specification of hESCs to hemogenic precursors? Second, does MLL-AF4 expression alter subsequent hematopoietic commitment of these hESC-derived hemogenic precursors? And, third, is enforced expression of MLL-AF4 in this cellular context sufficient to confer in vitro and/or in vivo proliferative or survival advantage as anticipated of a transforming oncogene?
In line with the well-established activation of clustered homeobox (Hox) genes by MLL fusions, our data show that MLL-AF4 also upregulates global Hox gene expression in hESCs. Functionally, MLL-AF4 influences the fate of hESCs and hESC-derived hemogenic precursors, as it first promotes the specification of hemogenic precursors from hESCs while later on it impairs further hematopoietic commitment of these precursors in favor of an endothelial cell fate. Importantly, MLL-AF4 expression is not sufficient to transform hESC-derived hematopoietic cells in vitro or in vivo. This work illustrates how hESCs can provide unique insights into human development and further our understanding of the early events regulating human embryonic hemato-endothelial specification.
Results
The expression of MLL-AF4 is compatible with hESC pluripotency and induces an activation pattern of Hox gene family expression
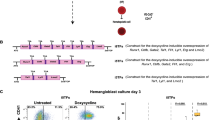
The leukemic fusion gene MLL-AF4 arises prenatally 5. To date, no study has explored the effects of leukemic fusion genes known to arise in utero during hESC-derived hematopoietic development. Here, MLL-AF4 cDNA was subcloned in a lentiviral vector expressing the Neomycin resistance cassette (NEO) (Figure 1A). Human ESCs were transduced with either the empty lentivector (NEO) or the MLL-AF4-expressing lentivector (MLL-AF4). After 3-4 weeks of G418 selection, typical neo-resistant hESC colonies emerged (Figure 1B). Successful and stable ectopic expression of MLL-AF4 in these hESCs was confirmed by RT-PCR (Figure 1C) and western blot (Figure 1D) more than 10 weeks after G418 selection. MLL-AF4 hESC cultures were then analyzed for pluripotency markers and functional assays. MLL-AF4-expressing hESCs retained high expression of both the pluripotency markers Oct4, Nanog and Rex-1 (Supplementary information, Figure S1A), and the hESC-associated antigens Tra-1-60, Tra-1-81, SSEA-3 and SSEA-4 (Supplementary information, Figure S1B). Functionally, MLL-AF4 and NEO hESCs formed teratomas with identical efficiency (100%), latency (50-65 days) and histological composition (Supplementary information, Figure S1C). Importantly, the ectopic expression of MLL-AF4 compares quite well between experimentally transduced cells and MLL-AF4-expressing leukemic cell lines (SEM and RS4;11) (Supplementary information, Figure S2).
Ectopic expression of MLL-AF4 in hESCs activates Hox gene expression. (A) Schematic representation of the lentiviral vectors used. (B) Phase contrast morphology of colonies from NEO- and MLL-AF4 hESCs. (C) RT-PCR confirming expression of the MLL-AF4 transcript in transduced hESCs. Line 1 was used as a housekeeping gene. (D) Western blot detection of the MLL-AF4 protein in transgenic hESCs. 293T cells transduced with MLL-AF4 and MLL-AF4ΔNH2 were used as positive and negative control, respectively. ERK1 and ERK2 blot was used as loading control. (E) Hox gene expression profiling in MLL-AF4 vs NEO hESC using microarrays. The inset shows qPCR validation of the indicated Hox genes/co-factors.
As MLL fusions are positive regulators of homeobox gene expression 18, 19, 20, we next performed microarray gene expression in NEO and MLL-AF4 hESCs to specifically analyze the impact of MLL-AF4 expression on the Hox gene family transcriptome in hESCs. As detailed in Figure 1E, 88.9% of the Hox family genes differentially expressed between MLL-AF4 and NEO hESCs resulted to be upregulated in MLL-AF4 hESCs, including the consistently upregulated targets of MLL fusions HOXA9 and MEIS1. Together, these data confirm successful expression of MLL-AF4 in hESCs that is compatible with pluripotency and activates Hox gene expression.
Augmented specification of hemogenic precursors from MLL-AF4 hESCs
We have established that the expression of MLL-AF4 is compatible with hESC pluripotency and upregulates global Hox gene expression. Next, we tested whether this prenatal leukemic fusion may impact the hemato-endothelial cell fate of hESCs using in vitro hematopoietic and endothelial differentiation from hESCs as surrogate assays for early developmental events. During human embryoid body (hEB) differentiation, a population of primitive hemogenic precursors arises, which is uniquely responsible for hematopoietic and endothelial development 14, 15. We thus investigated first the effect of MLL-AF4 on the emergence of hemogenic precursors (CD45-CD31+CD34+) throughout hEB development (Figure 2A and 2B). To ensure that any developmental effect is linked to MLL-AF4 expression, we confirmed stable transgene expression upon hEB differentiation by RT-PCR (Figure 2C). We found that the frequency of hemogenic precursors in MLL-AF4 hEBs was consistently higher (1.7-4.3-fold) throughout hEB development (Figure 2D). Importantly, MLL-AF4 expression accelerated the kinetics of emergence of hemogenic precursors. As shown in Figure 2D, by day 4 of hEB development, hemogenic precursors emerged in MLL-AF4 hESCs but barely did so in NEO hESCs.
MLL-AF4 promotes hESC specification towards hemogenic precursors but impairs subsequent hematopoietic differentiation of such hemogenic precursors. (A) Schematic of the hematopoietic differentiation of hESCs and endpoint analyses. (B) Representative flow cytometry dot plots displaying how hemogenic progenitors (CD45−CD31+), primitive blood cells (CD45+CD34+) and total blood cells (CD45+) are identified. (C) RT-PCR confirming stable MLL-AF4 expression upon differentiation (day 11 hEBs). (D) Specification into hemogenic precursors is significantly enhanced in MLL-AF4 hESCs throughout EB development. Subsequent differentiation of MLL-AF4 hemogenic precursors into primitive (E) and mature blood cells (F) is significantly decreased. (G) CFU read out from d15 and d22 hEBs confirming a reduced hematopoietic potential in MLL-AF4 cells. Data are presented as mean ± SEM for six independent experiments.
The increased frequency and accelerated emergence of hemogenic precursors in MLL-AF4 hESCs may be the result of either (i) MLL-AF4-mediated specification of hESCs towards hemogenic precursors or (ii) MLL-AF4-mediated enhanced proliferation of the emerging hemogenic precursors. To address this, cell cycle distribution and the proportion of sub-G0/G1 apoptotic cells was analyzed within both the hemogenic precursor population and the remaining EB cells (Supplementary information, Figure S3A). No differences in the proportion of cycling cells or apoptotic cells were observed either between MLL-AF4 and NEO hemogenic precursors (cycling cells: 25% vs 28%; apoptotic cells: 7% vs 12%) or between MLL-AF4 hemogenic precursors and the remaining MLL-AF4 EB cells (cycling cells: 25% vs 23%; apoptotic cells: 7% vs 10%) (Supplementary information, Figure S3A). Similarly, MLL-AF4 expression did not confer either growth or survival advantage to undifferentiated hESC cultures, as measured by BrdU incorporation (Supplementary information, Figure S3B). These data suggest that MLL-AF4 expression promotes specification, rather than selective proliferation, of hemogenic precursors from differentiating hEBs.
MLL-AF4 impairs hematopoietic commitment of hESC-derived hemogenic precursors
Hemogenic precursors derived from hESCs are uniquely responsible for endothelial and hematopoietic development 15, 21, 22. We next assessed whether the MLL-AF4-mediated enhanced specification of hemogenic precursors occurs with a subsequent increase of both hematopoietic and endothelial commitment. The emergence of primitive (CD45+CD34+) and total hematopoietic cells (CD45+) was analyzed throughout hEB development (Figure 2E and 2F). As expected, hematopoietic cells did not emerge prior to day 10 of hEB development (Figure 2E and 2F). Interestingly, the expression of MLL-AF4 abrogated the hematopoietic differentiation as reflected by a robust reduction on the frequency of both CD45+CD34+ (2.5- to 4.2-fold decrease) and total CD45+ cells (2-fold decrease) at day 11 and day 15 of hEB development (Figure 2E and 2F). Importantly, MLL-AF4 expression not only blocked the generation of CD45+ blood cells but also strongly compromised the clonogenic potential of hematopoietic progenitors derived from day 15 and day 22 hEBs (Figure 2G). Hematopoiesis generated from MLL-AF4 hESCs displayed a highly reduced (3-12-fold decrease) clonogenic potential measured by the ability to form colony-forming units (CFUs) in semisolid cultures (Figure 2G). Of note, MLL-AF4 effects were independent of the presence of hematopoietic cytokines, since an identical trend was observed in differentiation experiments carried out without BMP4 and hematopoietic growth factors (Supplementary information, Figure S4). These data indicate that the ectopic expression of MLL-AF4 promotes specification of hemogenic precursors from hESCs but impairs subsequent hematopoietic commitment of these precursors.
MLL-AF4 hemogenic precursors display enhanced endothelial cell fate
According to our results so far, MLL-AF4 expression promotes specification of hemogenic precursors from hESCs but impairs subsequent hematopoietic commitment of such precursors (Figure 2). Two distinct scenarios are plausible: the expression of MLL-AF4 in hemogenic precursors may (i) block both subsequent endothelial and hematopoietic differentiation or (ii) skew the hemato-endothelial commitment in favor of an endothelial cell fate. To explore these two possibilities, we analyzed the ability of both NEO and MLL-AF4 hemogenic precursors to differentiate into mature endothelial cells. NEO and MLL-AF4 hEBs were dissociated at day 11 of development and the hemogenic precursors were MACS-sorted and cultured for 5-7 days in conditions conductive to endothelial maturation (Figure 3A). After 5-7 days in endothelial culture conditions, the expression of the mature endothelial markers VE-cadherin, vWF and eNOS, and the uptake of LDL were examined in both NEO and MLL-AF4 cultures.
MLL-AF4 hemogenic precursors display an enhanced endothelial cell fate as compared to NEO hemogenic precursors. (A) Schematic of the endothelial differentiation and phenotypic and functional characterization of sorted hemogenic precursors. (B) NEO and MLL-AF4 hemogenic precursors isolated from day 11 EBs were cultured in EGM-2 media for 5-7 days and analyzed by immunohistochemistry for vWF, VE-cadherin and eNOS expression as well as LDL uptake (n = 3). Representative single stainings are shown on the right. A “merge” staining shows marker expression co-localization. (C) qPCR analysis of NEO and MLL-AF4 cells for the indicated endothelial-specific genes. This analysis was performed on NEO and MLL-AF4 hemogenic precursors after 6 days of culture in gelatin-coated plates, as indicated in A. (D) After 6 days of culture, NEO and MLL-AF4 hemogenic precursors were cultured in Matrigel at a density of 12 × 103 cells/well to assess their capacity to form capillary-like structures (n = 3). Images were captured 0 and 4 h after cell plating. Representative images are presented in the left panel. Quantification of the number of endothelial tubes, total length of tubes, and number of branching points was obtained with the software developed by Wimasis. Software-processed images are presented on the right as an example. Data are presented as mean ± SEM.
Both NEO and MLL-AF4 hemogenic precursors cultured in endothelial conditions became attached and spindle shaped. However, MLL-AF4 cells expressed stronger levels of VE-cadherin than NEO cells. More importantly, while NEO cells showed a predominant cytoplasmic localization for VE-cadherin, MLL-AF4 cells displayed a clear localization of VE-cadherin at the cell surface, identifying adherent junctions between cells, reminiscent of a distinctive endothelial barrier (Figure 3B). Similarly, the expression of vWF and eNOS was more robust in MLL-AF4 than in NEO endothelial cells (Figure 3B). In addition, MLL-AF4 endothelial cells also possessed higher LDL uptake capacity than NEO endothelial cells, also indicative of endothelial maturation (Figure 3B). A more quantitative assessment of this favored endothelial specification of MLL-AF4 hemogenic precursors was approached by quantitative PCR (qPCR) for VE-cadherin and vWF. MLL-AF4 endothelial cells consistently expressed 3-fold higher levels of both VE-cadherin and vWF than NEO endothelial cells (Figure 3C).
A key evidence of the acquisition of functional endothelial properties was evaluated by culturing NEO and MLL-AF4 hemogenic precursors on Matrigel to assess their capacity to form a network of capillary-like tubes. In line with the immunocytochemical and gene expression data, MLL-AF4 hemogenic precursors displayed an enhanced and faster ability than NEO precursors to align and form capillary-like structures (Figure 3D). As early as 4 h after seeding the cells in Matrigel, MLL-AF4 cultures, but not NEO cultures, contained capillary-like structures (Figure 3D, left panels). Computational quantification revealed that MLL-AF4 endothelial cells are more functional and mature than NEO endothelial cells since they displayed significantly higher number of capillary tubes, longer endothelial tubes and more branching points (Figure 3D, right panels). This data indicates that MLL-AF4 rather than blocking both endothelial and hematopoietic commitment of the hemogenic precursors, seems to skew the hemato-endothelial potential of these hemogenic precursors towards an endothelial cell fate.
Clonal analysis confirms that MLL-AF4 skews hemogenic precursor commitment towards endothelial cell fate
We next addressed whether MLL-AF4 skews the hemato-endothelial potential of the hemogenic precursors at the single cell level. Single hemogenic precursors isolated from day 11 NEO and MLL-AF4 hEBs were deposited into individual wells of 96-well plates and inspected daily. Individual wells targeted for NEO or MLL-AF4 single precursor cell deposition were visually inspected at 2 h and again after 16 h post-clonal isolation and only wells containing one cell were selected for further analysis (Figure 4A). This inspection was performed by two independent observers blinded to the other's results 15. Both observers demonstrated > 99% concordance of potential wells that contained more than one cell and these wells were then excluded from the subsequent analysis. After 12-15 days, the outgrowth in each well resulting from single hemogenic precursor proliferation was identified by phase contrast morphology (Figure 4B) and by DAPI staining (Figure 4C) and, analyzed in situ for expression of CD45 and VE-cadherin, representing hematopoietic and endothelial cell fate, respectively (Figure 4C) 15.
Clonal analysis confirms an enhanced endothelial commitment of MLL-AF4 hemogenic precursors. (A) Schematic of EB development and FACS sorting of single hemogenic precursors into 96-well plates. (B) Phase contrast images showing hematopoietic outgrowth only, endothelial outgrowth only or mixed (hemato-endothelial) outgrowth. Hematopoietic cells are identified as small round refractive cells loosely attached to the gelatin-coated plastic whereas endothelial cells are larger spindle-shaped cells strongly attached to the gelatin-coated plastic. (C) The cells resulting from single hemogenic precursor proliferation in each individual well were analyzed in situ for CD45 (hematopoietic cell fate) and VE-cadherin (endothelial cell fate). CD45 expression is shown in red, VE-cadherin in green and DAPI nucleus staining in blue. (D) Summary table illustrating the resulting outgrowth within individual wells containing single NEO or MLL-AF4 hemogenic precursors. At 10 to 15 days after sorting, single wells were observed to contain (i) hematopoietic progeny only, (ii) endothelial progeny only or (iii) both hematopoietic and endothelial cells.
Of the 936 and 960 wells containing, respectively, single NEO or MLL-AF4 hemogenic precursors, 44 wells (4.7%) and 41 wells (4.3%) demonstrated clonal outgrowth, respectively, consistent with the reported difficulty of sustaining single cells differentiated from ESCs in culture (Figure 4D) 15, 23. The resulting progeny of the single hemogenic precursor clones was then examined for hematopoietic and/or endothelial cell fate by in situ analysis of individual wells (Figure 4C). Consistent with the data from bulk cultures and hEB hematopoietic and endothelial differentiation (Figures 2 and 3), expression of MLL-AF4 strongly facilitated clonal endothelial over hematopoietic cell fate of single hemogenic precursors (Figure 4D). Progeny of NEO hemogenic precursor clones was exclusively hematopoietic in 23% of the wells (10 out of 44 wells) or exclusively endothelial in 54% of the wells (24 out of the 44 wells), while the remaining 10 clones (23% of the wells) were capable of giving rise to both endothelial and hematopoietic cells (Figure 4C and 4D). In sharp contrast, progeny of MLL-AF4 hemogenic precursors was exclusively endothelial in 97.5% of the clones (40 out of 41 wells) (Figure 4C and 4D) and no hematopoietic outgrowth from single MLL-AF4 hemogenic precursors could be detected. This clonal analysis confirms that MLL-AF4 expression robustly skews hemogenic precursor commitment towards endothelial cell fate.
MLL-AF4 acts as a global transcriptional activator and upregulates master genes associated to vascular-endothelial functions and early hematopoiesis
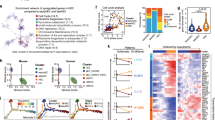
As we had performed microarray analysis of gene expression in undifferentiated NEO and MLL-AF4 hESCs, we looked back at our data in order to identify patterns of gene expression that could help explain at the molecular level the developmental effect of MLL-AF4 in hESCs. Global data analysis identified 1 267 genes differentially regulated (P value < 0.05; 2-fold regulation) between NEO and MLL-AF4 hESCs (Figure 5 and Supplementary information, Table S1). Overall, MLL-AF4 functioned as a transcriptional activator because its expression induced upregulation of 1 015 differentially expressed genes (80.1%) and downregulation of only 252 genes (19.9%) (Figure 5A). Analysis of the altered genes using the Ingenuity Analysis Program (IPA) software revealed that several gene functions (Figure 5B) and signaling pathways (Figure 5C) displayed a significantly altered gene expression profile in MLL-AF4 hESCs. Among the altered gene functions, tissue development, cancer, cardiovascular system development and function as well as hematopoietic system development are the most significantly upregulated cell functions upon the expression of MLL-AF4 in hESCs (Figure 5B and Supplementary information, Table S1). Similarly, among the altered signaling pathways, cardiogenesis, cardiomyocyte differentiation and VEGF signaling represent the most significantly upregulated signaling pathways in MLL-AF4 hESCs (Figure 5C and Supplementary information, Table S1).
Gene expression profiling and signaling pathways are altered in MLL-AF4 hESCs. (A) Heatmap showing a total of 1 267 genes differentially regulated between MLL-AF4 and NEO hESCs (P value < 0.05; 2-fold regulation). In all, 81% (1 015) of the genes differentially expressed are upregulated in MLL-AF4 hESCs. (B-E) After gene expression microarray analysis, the groups of genes differentially expressed (P value < 0.05; 2-fold regulation) in MLL-AF4 vs NEO hESCs were compared and the lists of functions and canonical pathways significantly altered were generated using the Ingenuity Pathways Analysis (IPA) 8 software. IPA software-based data mining generated a list of significantly modulated (up and down) gene functions (B) and canonical pathways (C) between MLL-AF4 and NEO hESCs. A more profound analysis was then performed for all the genes classified by the IPA software as involved in cardiovascular/vascular-endothelial system development and function (D) and hematopoietic system development and function (E). Master genes strongly associated with early hematopoiesis (i.e., SCL, RunX1, GATA2, CD34, BMP4, HOXB4) and vascular-endothelium (i.e., VEGFA, VEGFC, CD34, KDR, TIE1, TEK, SELE, NRP1, NRP2, COLs) likely to contribute to hemogenic precursor specification and subsequent endothelial development are underlined. Supplementary information, Table S1 contains the complete list of differentially regulated genes classified by the IPA software in the other gene functions and canonical pathways.
Those genes differentially upregulated in MLL-AF4 hESCs, classified by the IPA software as involved in cardiovascular/vascular-endothelial system development and function (Figure 5D) and hematopoietic system development and function (Figure 5E) were analyzed in more detail. We found upregulation of many key genes associated with early hematopoiesis including SCL/TAL1, RUNX1/AML1, GATA2, CD34, BMP4 and HOXB4 and with vascular-endothelium, such as VEGFA, VEGFC, CD34, KDR, TIE1, TEK, SELE, NRP1, NRP2 and different members of the collagen family. Supplementary information, Table S1 contains the complete list of differentially regulated genes classified by the IPA software belonging to other gene functions and canonical pathways. This gene expression profiling (GEP) indicates that MLL-AF4 hESCs seem transcriptionally primed to differentiate towards hemogenic precursors (hemangioblast) prone to subsequent endothelial maturation, as reflected by the marked upregulation of master genes associated to vascular-endothelial functions and early hematopoiesis.
Expression of MLL-AF4 is not sufficient for either in vitro or in vivo transformation of hESC-derived hematopoietic cells
Our findings so far support the idea that MLL-AF4 neither transforms undifferentiated hESC nor promotes hematopoiesis from hESCs. We next wanted to rule out the possibility that the expression of the MLL-AF4 specifically confers in vitro or in vivo proliferative and survival advantage to hESC-derived hematopoietic cells, as anticipated for a transforming leukemic oncogene. As shown in Figure 6A, MLL-AF4-expressing hESC-derived CD45+ hematopoietic cells did not display proliferative or survival advantage. Functionally, MLL-AF4 did not confer stable in vitro replating efficiency of hematopoietic cells in CFU assays (Figure 6B). Although hESC-derived hematopoietic cells have been extensively reported to barely repopulate immunodeficient mice, we finally attempted to ensure that MLL-AF4 does not affect the in vivo behavior of hEB-derived hematopoietic cells. Expectedly, MLL-AF4 did not confer in vivo function and engraftment potential to hESC-derived hematopoietic cells (Figure 6C), confirming that MLL-AF4 expression does not transform hESC-derived hematopoietic cells in vitro or in vivo, similar to that reported in neonatal CD34+ HSPCs 9.
MLL-AF4 does not transform hESC-derived CD45+ hematopoietic cells in vitro nor in vivo. MLL-AF4-expressing hESC-derived CD45+ hematopoietic cells do not show either proliferative/survival advantage (A) or stable in vitro replating efficiency in hematopoietic CFU assays (B) or in vivo hematopoietic engraftment potential into NSG mice (C).
Discussion
MLL-AF4+ pro-B ALL is a dismal infant leukemia that manifests in the first year of life 24. Mounting evidence indicates that MLL-AF4 is the initiating leukemogenic event with an in utero origin 5, 25. However, an understanding of potential changes in early hematopoietic development mediated by MLL-AF4 is lacking, despite of the recent advances by Krivtsov et al. 8 and Bursen et al. 26, current mouse models do not accurately recapitulate either the disease phenotype or latency 6, 8, 9. Furthermore, studies using primary cells from MLL-AF4 patients are incapable of addressing the developmental genesis of the hematopoietic system. The lack of bona fide putative MLL-AF4 oncogene models suggests that these approaches may be missing some essential aspects impacting hematopoietic cell fate during early human development. Postnatal (CB-derived) or prenatal (hESC-derived) stem cells represent ontogenically primitive target cells to address the developmental impact of MLL-AF4. Very recently, Montes et al. 9 explored for the first time the effect of MLL-AF4 on the fate of human neonatal HSPCs. MLL-AF4-expressing CB-HSPCs displayed an enhanced in vivo hematopoietic engraftment and in vitro clonogenic potential, but MLL-AF4 expression was not sufficient for leukemogenesis, indicating that either additional hits are required to develop leukemia or that CB-derived HSPCs do not represent the appropriate target for MLL-AF4 to induce transformation.
hESCs and hESC-derivatives enable the study of unique aspects of early human development that cannot otherwise be addressed by patient sample analyses or mouse models 11, 12. The fact that leukemogenesis manifests as a blockage or altered cell differentiation suggests that hematopoietic differentiation of hESCs could become a promising human-specific strategy to study the onset of hematopoiesis, particularly the emergence of the earliest events leading to the specification of the hematopoietic tissue 12. Previous studies made use of the BCR-ABL fusion gene to promote hematopoietic proliferation in mouse ESCs 27, 28 and only a very recent study has studied the impact of Nup98-HoxA10 fusion oncogene on hESC-derived hematopoiesis 29. However, this is the first study exploring the developmental impact of a leukemic fusion known to arise in utero on hESC hemato-endothelial development.
Despite being a leukemic oncogene, MLL-AF4 expression in hESCs or hESC-derived hematopoietic cells did not transform either in vitro or in vivo. In vitro, MLL-AF4+ hESC-derived hematopoietic cells did not display any proliferative and/or survival advantage and failed to confer replating efficiency in hematopoietic CFU assays, whereas in vivo, MLL-AF4 failed to endow hESC-derived hematopoiesis with engraftment potential upon xenotransplantation into NSG mice. This suggests that either additional hits or the reciprocal AF4-MLL 26 may be required for leukemogenesis, or that hESCs or hESC-derived hematopoietic derivatives are not the appropriate cellular targets for MLL-AF4-mediated transformation. It cannot be ruled out that other embryonic precursors (i.e., mesodermal precursors) or even fetal HSPCs may represent potential target cells in which MLL-AF4 originates and/or exerts its oncogenic function.
Importantly, MLL-AF4 did impact early hemato-endothelial specification from hESCs. It promoted the specification, rather than expansion, of early hemogenic precursors from differentiating hESCs. However, MLL-AF4 induced later developmental defects in the hematopoietic lineage as shown by a highly reduced production of both CD45+ and CD45+CD34+ hematopoietic cells and hematopoietic clonogenic potential. As the process of in vitro hematopoietic differentiation from hESCs closely mirrors early events in embryonic hematopoietic development, our results provide the first indication showing how the leukemic fusion gene MLL-AF4 impairs embryonic blood formation when experimentally overexpressed in hESCs. This is in agreement with the traditional view that hematopoietic dysfunction in newborn ALL carrying MLL-AF4 fusion might already occur at the embryonic/fetal stage, supporting the existence of concordant leukemia in monozygotic twins and suggesting that MLL-AF4 originates early in utero. Unexpectedly, MLL-AF4 expression led to an enhanced mature endothelial cell fate of the hemogenic precursors. Because hESC-derived hemogenic precursors are uniquely responsible for endothelial and hematopoietic development 15, 21, 22, 30, these data indicate that MLL-AF4 does not block both endothelial and hematopoietic commitment of the hemogenic precursors but instead, it skews the hemato-endothelial potential of these hemogenic precursors towards a pronounced endothelial cell fate.
The leukemia fusion oncogene BCR/ABL, as well as lymphoma-specific genetic aberrations, has been found in endothelial cells from chronic myeloid leukemia and B-cell lymphoma patients. This suggests that endothelial cells may be part of the neoplastic clone 31, 32 and that hemangioblasts/hemogenic precursors rather than HSPCs appear to be target cells for the first oncogenic hit, which could occur during the first steps of ESC differentiation and/or in hemangioblasts persisting in adults. In line with this, bone marrow-derived mesenchymal stem cells (BM-MSCs) from 100% infant pro-B ALL were recently found to harbor and express the MLL-AF4 fusion gene, suggesting that MLL-AF4 might arise in a population of pre-hematopoietic precursors 33. However, whether a potential endothelial dysfunction in MLL-AF4+ pro-B ALL patients exists remains to be clinically assessed. Interestingly, it has recently been demonstrated the existence of a common precursor of MSCs and endothelial cells, the mesenchymoangioblast, using hESCs directed towards mesendodermal differentiation 34. The existence of such a common embryonic precursor for MSCs and endothelium, and the reported expression of MLL-AF4 in both leukemic blasts and MSCs in the BM of infant patients suggest that MLL-AF4 fusion might arise and display a developmental impact on early pre-hematopoietic mesodermal or hemangioblastic precursors.
MLL fusions are well-known positive regulators of homeobox gene expression 18, 19. Interestingly, Stam et al. 19 have reported the existence of two distinct subgroups among t(4;11)-positive infant ALL cases characterized by the absence or presence of HOXA expression, with those patients lacking HOXA expression being at extreme high risk of disease relapse. In contrast, Trentin et al. 35 have recently questioned the function of activated Hox gene expression in t(4;11)-positive infant ALL. These patients were sub-divided into Hox gene expression high vs low and no biological/clinical relevance could be demonstrated. In line with this, MLL-AF4 expression in our experimental system robustly upregulated global Hox gene expression that was not oncogenic in a variety of assays, suggesting that activated Hox gene expression might be on its own not important for ALL.
Additionally, the enhanced specification of MLL-AF4 hESCs towards early hemogenic precursors (hemangioblasts) prone to subsequent endothelial maturation is reflected by gene expression analysis revealing a marked upregulation of key genes associated to neovasculature functions (VEGFA, VEGFC, CD34, KDR, TIE1, TEK, SELE, NRP1, NRP2 and several members of the collagen family) and early hematopoiesis (SCL/TAL1, RUNX1/AML1, GATA2, CD34, BMP4 and HOXB4), suggesting that MLL-AF4 hESCs seem transcriptionally primed to differentiate towards early hemogenic precursors as compared to NEO hESCs. Importantly, Guenther et al. 36 recently reported a list of 42 direct targets of MLL-AF4. Despite constituting different experimental approaches (non-transformed hESCs vs fully transformed leukemia cell lines), we have compared our GEP with the MLL-AF4 direct targets proposed by Guenther et al.36 and found that 8 (ERG, TNRC18, ADAM10, HOXA10, HOXA9, MEIS1, MEF2C, ZEB2) out of these 42 (20%) MLL-AF4 direct targets were also upregulated in our experimental system.
We have also compared our GEP with the GEP reported by Trentin et al. 35 and Stam et al. 19. As shown in Supplementary information, Figure 5SA, few genes were commonly found regulated not only among the three studies, but also between Trentin et al.35 and Stam et al. 19 (< 19 genes). Interestingly, Meis1, a key Hox co-factor implicated in leukemogenesis, was found upregulated in the three studies. The groups of genes differentially upregulated in the three studies were compared using IPA software. IPA software-based data mining generated a list of significantly upregulated gene functions and canonical pathways between MLL-AF4+ hESC/infant B-ALL and NEO hESCs/normal BM. As shown in Supplementary information, Figures 5SB, 5SC and S6, genes differentially upregulated in both MLL-AF4+ hESCs and MLL-AF4+ infant B-ALL were classified as involved in hematopoiesis, tissue development, VEFG signaling and cardiovascular/vascular-endothelial system development. It is crucial to be aware that these two GEP studies based on primary patient samples are conceptually distinct. Trentin et al.35 compared MLL-AF4+ infant B-ALL with normal bone marrow whereas Stam et al.19 compared MLL-AF4+ infant B-ALL with other MLL germline ALL, indicating that because gene expression data has been normalized against different primary samples, caution must be taken when comparing independent GEP studies. Additionally, caution is also required when comparing independent GEP studies mediated by MLL-AF4 because secondary oncogenic events already present in patient samples 19, 35 may contribute along with MLL-AF4 to the gene expression signatures observed in these clinical samples, whereas in our transgenic hESC model we studied the impact of gene expression of MLL-AF4 on its own (in a non-leukemic background).
Our experimental approach may constitute a new system to study the cellular and molecular mechanisms underlying MLL-AF4-mediated human early embryonic development. Because MLL-AF4 expression does not alter pluripotency, the unlimited replicative potential of hESCs enables the production of MLL-AF4-expressing embryonic hematopoietic, endothelial and MSC cells for studies that were previously unfeasible. Long-term, large-scale culture of MLL-AF4 hESC-derived hematopoietic cells or hemogenic precursors provides an unprecedented system for drug screening and toxicity studies. It also offers a unique in vitro system to test the ability of potential cooperating oncogenic events (reciprocal AF4-MLL or FLT3-activating mutations) or causal genotoxic compounds to induce leukemic transformation in vitro, characterized by the outgrowth of malignant B-lymphoid/monocyte clones 29, 37, 38. Taken together, our results provide the first indication showing how the leukemic fusion gene MLL-AF4 when overexpressed in hESCs impairs embryonic blood formation, establishing a potential novel experimental system to further study the developmental impact of MLL-AF4.
Materials and Methods
Plasmid construction and lentiviral transduction
The MLL-AF4 cDNA (MLL exon 10 was fused to AF4 exon 8; kindly provided by Professor Eric So, King's College London) was subcloned into the PmeI site of pRRL-EF1α-PGK-NEO vector (kindly provided by Professor L Naldini, ISR, Milano, Italy) 9. The following lentivectors were used: pRRL-EF1α-PGK-NEO (control; NEO) and pRRL-EF1α-MLLAF4-PGK-NEO (MLL-AF4). Viral particles pseudotyped with VSV-G were generated on 293T cells by standard calcium-phosphate transfection protocol and concentrated by ultracentrifugation as previously described 33. hESCs were infected overnight on the day of passage with concentrated virus in the presence of polybrene at 8 μg/ml (Sigma-Aldrich). At the following day, the viral supernanant was removed and infected hESCs were washed with fresh media and maintained in culture. After 3 days, transduced cells were selected with G418 (Invitrogen) at 50-100 μg/ml for 3 weeks. MLL-AF4 expression was confirmed in selected cells before being used for further experiments.
Human ESC culture
H9 and AND-1 hESCs were maintained undifferentiated in a feeder-free culture as previously described 14, 39. Briefly, hESCs were cultured in Matrigel (BD Biosciences, Bedford, MA, USA)-coated T25 flasks in human feeder conditioned medium (CM) supplemented with 8 ng/ml basic fibroblast growth factor (bFGF; Miltenyi, Germany) 40. Media was changed daily, and the cells were split weekly by dissociation with 200 U/ml of collagenase IV (Invitrogen, Edinburgh, Scotland). Human ESC cultures were visualized daily by phase contrast microscopy. Approval from the Spanish National Embryo Ethical Committee was obtained to work with hESCs.
RNA isolation, RT-PCR and qPCR analysis
Total RNA was isolated from undifferentiated hESCs, hEBs or hemogenic precursors using the Total RNA Purification Kit (Norgen, Canada) followed by RNase-free DNase treatment (Invitrogen). cDNA synthesis was done with 0.5 μg of total RNA using the First-Strand cDNA Synthesis Kit (Amersham, PA, USA). The resulting cDNA was used for conventional and qPCR. In qPCR experiments, the expression of each target gene was normalized to the expression of β-actin. qPCR was performed using power SYBR Green PCR Master Mix (Applied Biosystems) and the 7500 Real Time PCR System. Primer sequences used in this study are shown in Supplementary information, Table S2. For comparative expression of MLL-AF4 in transduced human stem cells and MLL-rearranged leukemic lines (SEM and RS4;11), the following primers (MLLAF4-Fw, 5′-CAGGTCCAGAGCAGAGCAAAC-3′ and MLLAF4-Rw, 5′-GAGCACTTGGAGGTGCAGATG-3′) and qRT-PCR conditions (95 °C for 10 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s) were used.
Flow cytometry characterization of hESCs
Flow cytometry analysis was carried out as previously described 41. Briefly, hESC cultures were dissociated with trypsin-EDTA and the single cell suspension was stained (2-5 × 105 cells/ml) with TRA-1-60-PE, TRA-1-81-FITC, SSEA3-PE and SSEA-4-FITC (all from BD Biosciences). The cells were then washed and stained with 7-AAD (BD Biosciences) for 15 min at room temperature. Cells were analyzed using a FACS Canto-II flow cytometer.
Western blot analysis
hESC cultures (3-5 × 105 cells) were dissociated with trypsin-EDTA for 10 min and the single cell suspension was subsequently lysed in 50 μl of RIPA buffer containing protease inhibitors for 30 min. 30 μl of the whole-cell lysate was mixed with 10 μl of loading buffer and resolved on 8% SDS-PAGE and transferred to PVDF membranes using a semi-dry transfer apparatus (at 15 V for 1 h). The membrane was blocked for 30 min with the blocking reagent BM Chemiluminiscence Western Blotting Kit (Roche). For MLL detection, the membrane was incubated overnight with anti-MLL antibody (1:100 dilution; clone N4.4; Millipore) followed by 2 h incubation with anti-mouse-HRP (Millipore). The MLL-AF4 fusion protein (250 KDa) was detected with the BM Chemiluminiscence Western Blotting Kit (Roche).
In vivo teratoma formation
Animal protocols were approved by the Local University Hospital Council On Animal Care and Experimentation. In vivo pluripotency was tested as previously described 42. Briefly, hESCs were harvested and implanted beneath the testicular capsule of 8-week-old immunodeficient male mice (The Jackson Lab, Bar Harbor, MA, USA). Teratoma growth was determined by palpation every week, and the mice were sacrificed ∼8-10 weeks after implantation. Teratomas were fixed, embedded in paraffin, and sections were stained with hematoxylin and eosin as described 42.
Hematopoietic differentiation from hESCs
Undifferentiated hESC cells at confluence were treated with collagenase IV and scraped off from the Matrigel attachments. They were then transferred to low-attachment plates (Corning, NY, USA) to allow hEB formation by overnight incubation in differentiation medium consisting of KO-DMEM supplemented with 20% fetal bovine serum (FBS), 1% nonessential amino acids, 1 mM L-glutamine, and 0.1 mM β-mercaptoethanol. Medium was changed the next day (day 1) with the same differentiation medium supplemented with hematopoietic cytokines: 300 ng/ml SCF, 300 ng/ml Flt-3L, 10 ng/ml IL-3, 10 ng/ml IL-6, 50 ng/ml G-CSF and 25 ng/ml BMP-4 14, 30, 37, 43. hEBs were dissociated using collagenase B (Roche Diagnostic, ON, Canada) for 2 h at 37 °C followed by 10 min incubation at 37 °C with Cell Dissociation Buffer (Invitrogen) at days 4, 7, 11 and 15 of development. A single cell suspension achieved by gentle pipetting and passaged through a 70-μm cell strainer was stained with anti-CD34-FITC, anti-CD31-PE, anti-CD45-APC (all from Miltenyi, Germany) and 7-AAD. Live cells identified by 7-AAD exclusion were analyzed using a FACS-Canto II flow cytometer (BD Biosciences). Hemogenic precursors with hemangioblastic properties were identified as CD34+CD31+CD45−. Immature and mature blood cells were identified as CD45+CD34+ and CD45+CD34−, respectively (Figure 2B) 14, 15, 30, 37.
Colony forming unit assay
Human clonogenic progenitor assays were performed by plating 50 000 cells from day 15 and day 22 hEBs into methylcellulose H4230 (Stem Cell Technologies, Vancouver, Canada) supplemented with recombinant human growth factors: 50 ng/ml SCF, 3 units/ml erythropoietin, 10 ng/ml GM-CSF and 10 ng/ml IL-3. Cells were incubated at 37 °C in a 5% CO2 humidified atmosphere and at day 14 colonies were counted with CFU assay using standard morphological criteria 14, 43, 44. For secondary re-plating, all the CFU colonies from each experimental condition (NEO and MLL-AF4) were harvested from the methylcellulose and a single cell suspension was achieved and re-plated as above 9.
Mice transplantation and analysis of engraftment
NOD/LtSz-scid IL-2Rγ−/− mice (NSG) were housed under sterile conditions. The Animal Care Committee of the University of Granada approved all mouse protocols. Briefly, CB-derived CD34+ HSPCs (n = 3, 3 × 104 cells in 50 μl) as well as NEO (n = 12) and MLL-AF4 (n = 15) day 15 hEB hematopoietic differentiating cells were transplanted intrahepatically (n = 20; 4-12 × 104 cells in 50 μl) into newborn NSG mice or intra-bone marrow (n = 7; 7-8 × 105 cells in 30 μl) as previously described in detail 45. Mouse health was monitored throughout the entire experiment. Mice were killed 6-8 weeks post-transplant and BM, spleen and liver were collected and analyzed for human chimerism. Cells from BM, spleen and liver were stained with anti-HLA-ABC-FITC, anti-CD31-PE and anti-CD45-APC (Beckton Dickinson) to analyze human chimerism by flow cytometry. Engrafted mice were assessed for multilineage analysis using anti-CD33-PE for myeloid cells, anti-CD19-APC for B-cells and anti-CD34-PE for immature hematopoietic cells (all from Miltenyi) 45.
GEP and data analysis
hESC samples were collected during the exponential cell growth phase and stabilized in RNAlater solution (Ambion, Austin, TX, USA) until RNA extraction. RNA was isolated using Agilent Total RNA Isolation Mini Kit (Agilent Technologies, Palo Alto, CA, USA) and its quality was checked in the Agilent 2100 Bioanalyzer platform. 500 ng of each total RNA sample was labeled with Cy3 using the Quick-Amp Labeling kit and hybridized with the Gene Expression Hybridization Kit to a Whole Human Genome Oligo Microarray (G4112F; 45 000 probes corresponding to 25 000 genes; Agilent Technologies) following the manufacturer's instructions. Each sample was labeled and hybridized as independent triplicates. Primary data was examined using GeneSpring 11.0 software (Silicon Genetics, Redwood City, CA, USA). A gene was considered differentially expressed if it was more than 2-fold regulated (up or down) in MLL-AF4 as compared to NEO hESCs. A t-test and a Benjamini Hochberg multi-testing correction were performed to better judge the significance of the regulated genes. Only genes satisfying the threshold of P value < 0.05 and a fold-change expression > 2 were included and assigned as significant. Pools of genes that were differentially expressed were clustered according to their expression pattern dynamics into hierarchical tree clustering algorithms using the Pearson's centered correlation distance definition as similarity measure and Centroid's as the linkage rule. Analysis of gene functions and canonical pathways significantly altered by MLL-AF4 was performed using the Ingenuity Pathway Analysis (IPA) software 8.0 (Ingenuity Systems, Inc., Redwood City, CA, USA). Several genes were confirmed by qPCR. Microarray data has been deposited and is available at Gene Expression Omnibus (GSE29869; http://www.ncbi.nlm.nih.gov/geo/).
Cell cycle analysis of hESCs, hemogenic precursors and CD45+ hematopoietic cells
Undifferentiated hESC cultures were stained with 10 μM BrdU (Sigma) for 20 min as previously described 9. The cells were then harvested, fixed in 70% ice-cold ethanol, and stored overnight at −20 °C. Subsequently, the cells were washed with PBS followed by 30 min incubation with 2 M HCl to depurinate the DNA. After washing with PBS containing 0.1% BSA and 0.2% Tween 20, the cells were incubated with 2 μl of anti-BrdU-FITC (BD Biosciences) for 30 min. After washing, the cells were resuspended in PI buffer containing 5 μg of PI and 100 μg/ml of RNAase in PBS.
For cell cycle analysis of hESC-derived hemogenic precursors and CD45+ hematopoietic cells, the hEBs were dissociated at day 11 and day 22 of development, respectively. Cells were then washed with PBS and incubated with anti-CD31-FITC and anti-CD34-FITC or anti-CD45-FITC, respectively, for 15 min. After washing, the cells were suspended in PI buffer as described above. BrdU staining of undifferentiated hESCs and cell cycle distribution of hemogenic precursors was analyzed on a FACS Canto-II cytometer discriminating among apoptotic cells (Sub-G0/G1), quiescent cells (G0/G1), cycling cells (S-phase, BrdU+) and G2/M cells 46, 47. The apoptotic status of NEO and MLL-AF4 hESC-derived CD45+ hematopoietic cells was assessed using the Annexin-V apoptosis detection kit (BD Biosciences), according to the manufacturer's instructions 9. Briefly, day 22 EBs were dissociated and washed twice with cold PBS before staining with anti-CD45-FITC, Annexin-V-PE and 7-AAD. Apoptotic cells were detected by gating the Annexin V+/7-AAD+ fraction.
Endothelial differentiation from hESCs, uptake of acetylated LDL and in situ immunocytochemistry
CD34+ cells were isolated from hEBs at day 11 of development by magnetic-activated cell sorting (MACS) using the hCD34 MicroBead kit and the AutoMACS Pro separator (Miltenyi Biotech), as per manufacturer's instructions 37, 46. To promote endothelial differentiation, isolated CD34+ cells were seeded on 0.1% gelatin-coated plates at 1.2 × 104 cells/cm2 in complete EGM-2 media (Lonza, Walkersville) for 5-7 days. For uptake of acetylated LDL, cells were incubated with 10 μg/ml of Dil-Ac-LDL (Molecular Probes) at 37 °C for 4 h. After fixation and permeabilization, cells were stained with rabbit anti-human VE-cadherin (Cayman, MI, FL, USA), mouse anti-human eNOS (BD Biosciences), mouse anti-human vWF (DAKO) and mouse anti-human CD45 (BD Biosciences) followed by FITC-conjugated anti-rabbitt or Cy3-conjugated anti-mouse antibodies (Jakson Immunoresearch), respectively.
For capillary-like tube formation assay, MACS-isolated CD34+ cells were expanded for 5-7 days in complete EGM-2 media and 1.2 × 104 cells/well were then seeded on top of 25 μl of Matrigel previously spreaded onto a 96-well plate. Pictures were captured (5× objective) at the indicated times with an Axiocam MRM digital camera (Zeiss) attached to an AxioImager A1 microscope (Zeiss). For quantification, all the images were analyzed with the WimTube software (www.wimasis.com; Wimasis SL, Munich, Germany). This object recognition tool identifies cellular tubes, branching points and loops automatically without manual adaptation.
Clonal experiments with FACS-sorted single hemogenic precursors
Single CD34+CD31+CD45− hemogenic precursors FACS-isolated from day 11 NEO hEBs or MLL-AF4 hEBs were deposited into individual wells of 96-well plates and inspected daily. Ten 96-well plates (∼960 single cells) were used per condition. Single cell sorting was carried out using a FACSAria sorter equipped with an Automatic Cell Deposition Unit (ACDU). Sorted single cells were allowed to expand and differentiate in 200 μl of a 50:50 mixture of hematopoietic (100 μl of StemSpam supplemented with BMP4, SCF, FLT3 and IL3) and endothelial (100 μl of complete EGM-2 media) supportive media. After 12-15 days, the resulting clonal outgrowth in each well was analyzed in situ by phase contrast morphology and immunocytochemical staining for CD45 (hematopoietic) and VE-cadherin (endothelial). The nuclei were counterstained with DAPI 15.
References
Meyer C, Schneider B, Jakob S, et al. The MLL recombinome of acute leukemias. Leukemia 2006; 20:777–784.
Caslini C, Alarcon AS, Hess JL, Tanaka R, Murti KG, Biondi A . The amino terminus targets the mixed lineage leukemia (MLL) protein to the nucleolus, nuclear matrix and mitotic chromosomal scaffolds. Leukemia 2000; 14:1898–1908.
Pui CH . Acute lymphoblastic leukemia in children. Curr Opin Oncol 2000; 12:3–12.
Greaves MF, Maia AT, Wiemels JL, Ford AM . Leukemia in twins: lessons in natural history. Blood 2003; 102:2321–2333.
Ford AM, Ridge SA, Cabrera ME, et al. In utero rearrangements in the trithorax-related oncogene in infant leukaemias. Nature 1993; 363:358–360.
Chen W, Li Q, Hudson WA, Kumar A, Kirchhof N, Kersey JH . A murine Mll-AF4 knock-in model results in lymphoid and myeloid deregulation and hematologic malignancy. Blood 2006; 108:669–677.
Metzler M, Forster A, Pannell R, et al. A conditional model of MLL-AF4 B-cell tumourigenesis using invertor technology. Oncogene 2006; 25:3093–3103.
Krivtsov AV, Feng Z, Lemieux ME, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell 2008; 14:355–368.
Montes R, Ayllon V, Gutierrez-Aranda I, et al. Enforced expression of MLL-AF4 fusion in cord blood CD34+ cells enhances the hematopoietic repopulating cell function and clonogenic potential but is not sufficient to initiate leukemia. Blood 2011; 117:4746–4758.
Menendez P, Bueno C, Wang L, Bhatia M . Human embryonic stem cells: potential tool for achieving immunotolerance? Stem Cell Rev 2005; 1:151–158.
Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science 1998; 282:1145–1147.
Lensch MW, Daley GQ . Scientific and clinical opportunities for modeling blood disorders with embryonic stem cells. Blood 2006; 107:2605–2612.
Menendez P, Vargas A, Bueno C, et al. Quantitative analysis of bcl-2 expression in normal and leukemic human B-cell differentiation. Leukemia 2004; 18:491–498.
Ramos-Mejia V, Melen GJ, Sanchez L, et al. Nodal/Activin signaling predicts human pluripotent stem cell lines prone to differentiate towards the hematopoietic lineage. Mol Ther 2010; 18:2173–2181.
Wang L, Li L, Shojaei F, et al. Endothelial and hematopoietic cell fate of human embryonic stem cells originates from primitive endothelium with hemangioblastic properties. Immunity 2004; 21:31–41.
Diehl F, Rossig L, Zeiher AM, Dimmeler S, Urbich C . The histone methyltransferase MLL is an upstream regulator of endothelial-cell sprout formation. Blood 2007; 109:1472–1478.
Hatzipantelis ES, Athanassiou-Metaxa M, Gombakis N, et al. Thrombomodulin and von Willebrand factor: relation to endothelial dysfunction and disease outcome in children with acute lymphoblastic leukemia. Acta Haematol 2011; 125:130–135.
Bach C, Buhl S, Mueller D, Garcia-Cuellar MP, Maethner E, Slany RK . Leukemogenic transformation by HOXA cluster genes. Blood 2010; 115:2910–2918.
Stam RW, Schneider P, Hagelstein JA, et al. Gene expression profiling-based dissection of MLL translocated and MLL germline acute lymphoblastic leukemia in infants. Blood 2010; 115:2835–2844.
Tkachuk DC, Kohler S, Cleary ML . Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 1992; 71:691–700.
Vodyanik MA, Bork JA, Thomson JA, Slukvin II . Human embryonic stem cell-derived CD34+ cells: efficient production in the coculture with OP9 stromal cells and analysis of lymphohematopoietic potential. Blood 2005; 105:617–626.
Vodyanik MA, Thomson JA, Slukvin II . Leukosialin (CD43) defines hematopoietic progenitors in human embryonic stem cell differentiation cultures. Blood 2006; 108:2095–2105.
Lumelsky N, Blondel O, Laeng P, Velasco I, Ravin R, McKay R . Differentiation of embryonic stem cells to insulin-secreting structures similar to pancreatic islets. Science 2001; 292:1389–1394.
Pui CH, Carroll WL, Meshinchi S, Arceci RJ . Biology, risk stratification, and therapy of pediatric acute leukemias: an update. J Clin Oncol 2011; 29:551–565.
Bueno C, Montes R, Catalina P, Rodriguez R, Menendez P . Insights into the cellular origin and etiology of the infant pro-B acute lymphoblastic leukemia with MLL-AF4 rearrangement. Leukemia 2011; 25:400–410.
Bursen A, Schwabe K, Ruster B, et al. The AF4.MLL fusion protein is capable of inducing ALL in mice without requirement of MLL.AF4. Blood 2010; 115:3570–3579.
Perlingeiro RC, Kyba M, Daley GQ . Clonal analysis of differentiating embryonic stem cells reveals a hematopoietic progenitor with primitive erythroid and adult lymphoid-myeloid potential. Development 2001; 128:4597–4604.
Peters DG, Klucher KM, Perlingeiro RC, Dessain SK, Koh EY, Daley GQ . Autocrine and paracrine effects of an ES-cell derived, BCR/ABL-transformed hematopoietic cell line that induces leukemia in mice. Oncogene 2001; 20:2636–2646.
Ji J, Risueno RM, Hong S, et al. Brief report: ectopic expression of Nup98-HoxA10 augments erythroid differentiation of human embryonic stem cells. Stem Cells 2011; 29:736–741.
Wang L, Menendez P, Shojaei F, et al. Generation of hematopoietic repopulating cells from human embryonic stem cells independent of ectopic HOXB4 expression. J Exp Med 2005; 201:1603–1614.
Fang B, Zheng C, Liao L, et al. Identification of human chronic myelogenous leukemia progenitor cells with hemangioblastic characteristics. Blood 2005; 105:2733–2740.
Streubel B, Chott A, Huber D, et al. Lymphoma-specific genetic aberrations in microvascular endothelial cells in B-cell lymphomas. N Engl J Med 2004; 351:250–259.
Menendez P, Catalina P, Rodriguez R, et al. Bone marrow mesenchymal stem cells from infants with MLL-AF4+ acute leukemia harbor and express the MLL-AF4 fusion gene. J Exp Med 2009; 206:3131–3141.
Vodyanik MA, Yu J, Zhang X, et al. A mesoderm-derived precursor for mesenchymal stem and endothelial cells. Cell Stem Cell 2010; 7:718–729.
Trentin L, Giordan M, Dingermann T, Basso G, Te Kronnie G, Marschalek R . Two independent gene signatures in pediatric t(4;11) acute lymphoblastic leukemia patients. Eur J Haematol 2009; 83:406–419.
Guenther MG, Lawton LN, Rozovskaia T, et al. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev 2008; 22:3403–3408.
Bueno C, Catalina P, Melen GJ, et al. Etoposide induces MLL rearrangements and other chromosomal abnormalities in human embryonic stem cells. Carcinogenesis 2009; 30:1628–1637.
Ono R, Nakajima H, Ozaki K, et al. Dimerization of MLL fusion proteins and FLT3 activation synergize to induce multiple-lineage leukemogenesis. J Clin Invest 2005; 115:919–929.
Cortes JL, Sanchez L, Ligero G, et al. Mesenchymal stem cells facilitate the derivation of human embryonic stem cells from cryopreserved poor-quality embryos. Hum Reprod 2009; 24:1844–1851.
Montes R, Ligero G, Sanchez L, et al. Feeder-free maintenance of hESCs in mesenchymal stem cell-conditioned media: distinct requirements for TGF-beta and IGF-II. Cell Res 2009; 19:698–709.
Menendez P, Perez-Simon JA, Mateos MV, et al. Influence of the different CD34+ and CD34− cell subsets infused on clinical outcome after non-myeloablative allogeneic peripheral blood transplantation from human leucocyte antigen-identical sibling donors. Br J Haematol 2002; 119:135–143.
Gutierrez-Aranda I, Ramos-Mejia V, Bueno C, et al. Human induced pluripotent stem cells develop teratoma more efficiently and faster than human embryonic stem cells regardless the site of injection. Stem Cells 2010; 28:1568–1570.
Chadwick K, Wang L, Li L, et al. Cytokines and BMP-4 promote hematopoietic differentiation of human embryonic stem cells. Blood 2003; 102:906–915.
Menendez P, Prosper F, Bueno C, et al. Sequential analysis of CD34+ and CD34− cell subsets in peripheral blood and leukapheresis products from breast cancer patients mobilized with SCF plus G-CSF and cyclophosphamide. Leukemia 2001; 15:430–439.
Bueno C, Montes R, de la Cueva T, Gutierrez-Aranda I, Menendez P . Intra-bone marrow transplantation of human CD34(+) cells into NOD/LtSz-scid IL-2rgamma(null) mice permits multilineage engraftment without previous irradiation. Cytotherapy 2010; 12:45–49.
Bueno C, Montes R, Menendez P . The ROCK inhibitor Y-27632 negatively affects the expansion/survival of both fresh and cryopreserved cord blood-derived CD34+ hematopoietic progenitor cells. Stem Cell Rev 2010; 6:215–223.
Rubio R, Garcia-Castro J, Gutierrez-Aranda I, et al. Deficiency in p53 but not Retinoblastoma induces the transformation of mesenchymal stem cells in vitro and initiates Leiomyosarcoma in vivo. Cancer Res 2010; 70:4185–4194.
Acknowledgements
This work was funded by the CICE/FEDER (P08-CTS-3678) de la Junta de Andalucía to PM, the FIS/FEDER (PI10/00449) to PM, (PI11/00119) to CB and (PS09/02454) to MFF, the ISCIII (CP07/00059) to CB, (CP09/0063) to PJR and (EMER07/055 and PI10/00883) to JC R-M, the MICINN (PLE-2009-0111) to PM, the Foundation “Spanish Association Against Cancer”/Junta Provincial de Albacete (CI110023) to PM, the Marie Curie IIF (PIIF-GA-2009-236430) to V R-M, the CSIC (200820I172) to MFF, and the Community of Asturias (FICYT IB09-106) to MFF. AFF is supported by the IUOPA-Obra Social Cajastur.
Author information
Authors and Affiliations
Corresponding authors
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
Enforced expression of MLL-AF4 is compatible with hESC pluripotency. (PDF 138 kb)
Supplementary information, Figure S2
Q-RT-PCR comparing the expression levels of MLL-AF4 in MLL-AF4-carrying leukemic cell lines and MLL-AF4-transduced human stem cells. (PDF 37 kb)
Supplementary information, Figure S3
Cell cycle analysis reveals that the expression of MLL-AF4 promotes specification of hESCs into hemogenic precursors rather than enhanced proliferation or survival of the emerging hemogenic precursors. (PDF 62 kb)
Supplementary information, Figure S4
Spontaneous hEB differentiation reveals that MLL-AF4 augments hemogenic precursor specification and impairs further hematopoietic commitment regardless the presence of hematopoietic cytokines. (PDF 25 kb)
Supplementary information, Figure S5
Comparative analysis of our gene expression profiling (GEP) with the GEP reported by Trentin et al. (2009) and Stam et al. (2010). (PDF 100 kb)
Supplementary information, Figure S6
mRNA expression of FLT3 (A-C) and wild-type MLL (D-F) in NEO- and MLL-AF4-expressing hESCs and day 15 hEBs. (PDF 62 kb)
Supplementary information, Table S1
Annotations and analysis of the differentially regulated genes between NEO and MLL-AF4 hESCs classified by the IPA software in relevant gene functions and canonical pathways. (XLS 684 kb)
Supplementary information, Table S2
Primer sequences used in the present study (PDF 12 kb)
Rights and permissions
About this article
Cite this article
Bueno, C., Montes, R., Melen, G. et al. A human ESC model for MLL-AF4 leukemic fusion gene reveals an impaired early hematopoietic-endothelial specification. Cell Res 22, 986–1002 (2012). https://doi.org/10.1038/cr.2012.4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2012.4
Keywords
This article is cited by
-
Panobinostat (LBH589) increase survival in adult xenografic model of acute lymphoblastic leukemia with t(4;11) but promotes antagonistic effects in combination with MTX and 6MP
Medical Oncology (2022)
-
Inhibition of DOT1L and PRMT5 promote synergistic anti-tumor activity in a human MLL leukemia model induced by CRISPR/Cas9
Oncogene (2019)
-
Molecular processes involved in B cell acute lymphoblastic leukaemia
Cellular and Molecular Life Sciences (2018)
-
NG2 antigen is involved in leukemia invasiveness and central nervous system infiltration in MLL-rearranged infant B-ALL
Leukemia (2018)
-
Reprogramming human B cells into induced pluripotent stem cells and its enhancement by C/EBPα
Leukemia (2016)