Abstract
The nuclear receptor PPARs are fundamentally important for energy homeostasis. Through their distinct yet overlapping functions and tissue distribution, the PPARs regulate many aspects of energy metabolism at the transcriptional level. Functional impairment or dysregulation of these receptors leads to a variety of metabolic diseases, while their ligands offer many metabolic benefits. Studies of these receptors have advanced our knowledge of the transcriptional basis of energy metabolism and helped us understand the pathogenic mechanisms of metabolic syndrome.
Similar content being viewed by others
Introduction
The prevalence of obesity has increased at an alarming rate in the last several decades worldwide. Along with it comes the associated metabolic syndrome, including insulin resistance, glucose intolerance, type 2 diabetes, dyslipidemia (increased serum triglycerides, decreased high-density lipoproteins (HDLs), and increased low-density lipoproteins (LDLs)), hypertension, nonalcoholic fatty liver, and cardiovascular disease 1. At the center of this medical cluster is insulin resistance, which is often caused by obesity. Insulin resistance not only is an early, fundamental defect in type 2 diabetes 2, but also is the defining feature of metabolic syndrome 1. Insulin resistance is manifested by hyperinsulinemia, decreased insulin-stimulated glucose uptake into skeletal muscle, and the impaired ability of insulin to inhibit gluconeogenesis in liver and lipolysis in adipose tissue. Type 2 diabetes occurs when the pancreatic β cells are unable to secrete enough insulin to compensate for insulin resistance. Although most insulin-resistant individuals are able to maintain relatively normal glucose levels through hyperinsulinemia during their lifetime, insulin resistance puts these individuals at great risk for the development of a variety of abnormalities associated with metabolic syndrome 1. Thus, obesity and insulin resistance play a central role in metabolic syndrome.
In order to better understand the functions of the nuclear receptor PPARs in energy metabolism and metabolic syndrome, it is useful to first summarize the potential mechanisms that could explain the pathogenesis of insulin resistance. Currently two theories are widely accepted. The first one hypothesizes that insulin resistance is tightly linked to disordered fatty acid metabolism and that insulin resistance results from the deleterious effects of lipid overload in the skeletal muscle and liver 3. Fatty acids are preferably stored as triglycerides in the adipose tissue. However, a dysregulation of fatty acid metabolism can lead to abnormal triglyceride accumulation in skeletal muscle and/or liver that in most cases associates with insulin resistance. This close association between insulin resistance and no-adipose lipid accumulation has been found not only in many animal genetic models, but also in human populations that include obese subjects, lipodystrophic patients, the lean offspring of type 2 diabetes, and the elderly 3. Interestingly, it is the increased intermediate metabolites of triglycerides, not triglycerides per se, that may be responsible for insulin resistance 3. One such metabolite is diacylglycerol. It is thought that diacylglycerol activates members of protein kinase PKC, which leads to serine phosphorylation of insulin receptor substrate-1 (IRS-1) and IRS-2 in the skeletal muscle and liver, respectively, thereby interfering with insulin action 3. The second theory links obesity-associated insulin resistance to inflammation. Obesity is considered as a chronic, low-grade inflammatory state. Liver and, in particular, adipose tissue are the sites for this inflammatory response. Obesity leads to an increased production of proinflammatory cytokines by adipose tissue, liver, and their recruited macrophages. These cytokines in turn inhibit insulin signaling both locally and systematically 4, 5. These two potential mechanisms are not necessarily mutually exclusive. Their relative importance for insulin resistance may depend on the pathophysiological states and affected tissues. Interestingly, PPARs appear to be at the crossroads of lipid metabolism and inflammation, regulating both processes 6.
The PPAR subfamily of nuclear receptors, consisting of three members, PPARα, PPARγ and PPARδ (also known as PPARβ), are ligand-activated transcription factors 6, 7. They form obligate heterodimers with retinoid X receptors and bind to specific DNA sites composed of a direct repeat of hexameric sequences separated by one base pair, located in the promoter/enhancer region of target genes. Ligand binding triggers a conformation shift that results in release of co-repressors, recruitment of co-activators, and subsequent target gene expression. As nuclear receptors, the PPARs have a typical domain organization, containing an amino-terminal domain, a central zinc-finger DNA-binding domain (DBD), a carboxyl-terminal ligand-binding domain (LBD) that harbors the ligand-dependent activation function, and a small hinge region that links DBD and LBD. The DBD and LBD are highly conserved among the three PPARs. Different from other nuclear receptors, the ligand-binding pocket of the PPARs is unusually large and can accommodate a variety of endogenous lipids, including fatty acids, eicosanoids, oxidized and nitrated fatty acids, and derivatives of linoleic acid 6. However, these lipids need to be present at high concentrations (micromolar range) to activate PPARs, and are often not highly selective for individual isoforms. Moreover, it has not been shown that any of these lipids is required for PPAR activation. Thus, whether any of them represents physiological ligand remains to be established. Nevertheless, it is clear that the activities and/or expression levels of PPARs are subjected to modulation by diets, nutrient status, and metabolic states. Through their distinct yet overlapping functions and tissue distribution, the three PPARs act as fatty acid sensors to control many metabolic programs that are essential for systematic energy homeostasis. Not surprisingly, they have important implications for human metabolic diseases, as evidenced by the fact that PPARγ and PPARα are respective molecular targets for the type 2 diabetes drug thiazolidinediones (TZDs) and dyslipidemia drug fibrates. Here I review our current understanding of these receptors in lipid metabolism and associated metabolic syndrome. For the roles of these receptors in inflammatory response that also play a part in metabolic syndrome, readers are referred to recent publications 6, 8, 9.
PPARγ is a master regulator of adipogenesis
Adipose tissue is essential for whole body energy homeostasis 10, 11. It has two functionally distinct types, white fat (WAT) and brown fat (BAT). WAT serves as a safe place that stores excess energy to avoid lipid build-up in other tissues, and releases the energy when other tissues are in need. It also has an endocrinal function, producing many secreted factors called adipokines that signal whole body metabolism 10, 11. A large body of work has clearly established that, through its induction of many genes important for fatty acid uptake and storage, PPARγ is both sufficient and necessary for the differentiation of white fat adipocytes 12, 13. PPARγ is predominantly expressed in the adipose tissue compared to other tissues and is induced during adipocyte differentiation 14. Ectopic expression of PPARγ in nonadipogenic cells effectively converts them into adipocytes 15, whereas knockout of PPARγ in embryonic fibroblasts abolishes their differentiation into adipocytes 16. Studies with germline deletion of PPARγ as well as studies with PPARγ hypomorphic mice have demonstrated an essential requirement for this gene in the formation of adipose tissue in vivo; that is, animals cannot generate adipocytes in the absence of PPARγ 17, 18, 19. Moreover, mice with PPARγ deleted in differentiated adipocytes develop progressive lipodystrophy, revealing that PPARγ is also important for the survival of differentiated adipocytes 20, 21, 22. Finally, heterozygous, dominant-negative PPARγ mutations cause lipodystrophy in humans 23.
PPARγ-controlled differentiation of white fat adipocytes involves a transcriptional cascade that includes members of C/EBP transcription factors 12, 13 (Figure 1A). In vivo studies suggest that C/EBPβ, δ and α are critical for adipogenesis 24, 25. How do the C/EBPs regulate adipogenesis? Largely based on in vitro differentiation data, the current prevalent view is that the three C/EBPs work in concert to induce and maintain PPARγ expression 12, 13. In addition, C/EBPα appears to be required for the insulin-sensitive properties of adipocytes 26, 27. Recently, it was observed that C/EBPα co-occupies many of the PPARγ-target gene promoters 28, raising the possibility that C/EBPα may also directly regulate adipogenesis in a cooperative manner with PPARγ. PTIP is a transcriptional co-factor that regulates the expression of C/EBPα and PPARγ through histone methylation of their promoters 29. The observation that rescue of the adipogenic defect in PTIP-deficient cells requires both PPARγ and C/EBPα is in agreement with this possibility 29. In vitro studies have identified additional transcriptional regulators in adipogenesis, including Med23, ELK1, and Krox20, which act upstream of C/EBPβ 30, 31, and KLF5 and XBP1, which act downstream of C/EBPβ 32, 33 (Figure 1A); however, the in vivo requirements for these factors in adipose tissue formation are unclear.
As mentioned above, there are two types of adipose tissue. In contrast to the energy storage function of WAT, BAT is specialized for energy expenditure by dissipating energy as heat 34, 35. The unique metabolic property of BAT is due to its high mitochondrial density and fuel oxidation capacity, and exclusive expression of the BAT hallmarker, uncoupling protein-1 (UCP1), in the inner mitochondrial membrane. UCP1 is a proton transporter that disrupts the mitochondrial electrochemical gradient by causing protons to leak across the inner mitochondrial membrane. Thus, UCP1 uncouples mitochondrial respiration from ATP production, leading to energy loss in the form of heat. BAT is located in the interscapular region in small mammals. In rodents, it is clear that BAT plays an important part in protection against obesity and obesity-associated metabolic problems. Previously, it was thought that BAT is only present in human neonates and is replaced by WAT in adults. However, recent work has collectively demonstrated that humans do possess functional brown fat depots and that the activity of BAT is inversely associated with obesity 36, 37, 38, suggesting that BAT could be important for human physiology and metabolic diseases.
There is a great interest in understanding the development of brown fat. Surprisingly, lineage studies show that BAT and WAT do not share the same precursor cells in early development; instead, BAT originates from Myf5-positive precursor cells that also give rise to skeletal muscle cells 39. As in WAT, PPARγ is highly expressed in BAT and is required for BAT formation 18. However, PPARγ alone generates a fat phenotype that is common to both WAT and BAT. The questions are what controls PPARγ expression in the BAT and what determines BAT-specific programs such as UCP1 expression and mitochondrial biogenesis. Spiegelman and co-workers discovered that C/EBPβ and transcriptional co-activators PRDM16, PGC-1α and PGC-1β are critical 39, 40, 42, 43. All these factors are selectively expressed in BAT compared to WAT. During brown fat cell differentiation, PRDM16 forms a complex with C/EBPβ and co-activates C/EBPβ transcriptional activity 42. This PRDM16/C/EBPβ complex not only induces PPARγ expression, but also induces PGC-1α expression, which is further activated by PRDM16 and is responsible for the BAT-specific program 39, 40, 42 (Figure 1B). Indeed, expression of either PGC-1α or PRDM16 in white fat preadipocytes stimulates the expression of BAT-specific genes during differentiation 40, 41, while brown fat preadipocytes deficient for PRDM16 or PGC-1α and PGC-1β lose differentiation-associated BAT-specific gene expression 40, 43. Moreover, co-expression of C/EBPβ and PRDM16 is sufficient to convert skin fibroblasts into functional brown fat adipocytes 42. These pioneering studies have significantly advanced our understanding of brown fat adipogenesis. While the physiological requirement for C/EBPβ in brown fat development is clear 42, can forced expression of C/EBPα or δ functionally substitute for C/EBPβ? In addition to C/EBPβ, are there other transcription factors mediating the induction of PGC-1α by PRDM16 during adipogenesis? As PPARγ can be co-activated by PRDM16 and PGC-1, does PPARγ directly regulate the expression of some brown fat-specific genes through these co-activators? Interestingly, it was shown that PPARγ is not required for UCP1 expression 40. Finally, other factors, including Rb and p107 44, RIP140 45, 46, and BMP7 47, have been identified as brown fat differentiation regulators. Their relationships with the molecular pathway described above remain to be determined.
PPARγ and TZD in insulin resistance and the mechanism of action
Members of TZD class drugs are widely used in the treatment of type 2 diabetes. In both animal models and human subjects with type 2 diabetes, TZDs lower glucose levels as well as insulin levels. Glucose clamp experiments demonstrate that TZDs increase skeletal muscle glucose disposal and, to a lesser extent, inhibit hepatic gluconeogenesis. Thus, TZDs work as insulin sensitizers by improving insulin action in the skeletal muscle and liver 48. However, clinical use of TZDs is associated with side effects, including weight gain, fluid retention, edema, and heart failure 49. How to separate the side effects from its efficacy is an important task in the future.
TZD drugs and PPARγ were independently discovered. The initial link between them was provided by findings that TZDs are in fact agonists for PPARγ 50, 51. It is now clear that activation of PPARγ by TZDs is responsible for their beneficial effects on insulin sensitivity. How does activation of PPARγ, a protein that is mainly present in adipose tissue, lead to insulin sensitization in skeletal muscle and liver? Activation of PPARγ in the adipose tissue induces expression of an array of genes for fatty acid transport and storage, as well as promotes de novo adipogenesis. One plausible mechanism is that the increased ability to uptake lipids and the expanded capacity to store them allow lipid repartitioning from the skeletal muscle and liver to the adipose tissue, thus eliminating the deleterious effects of lipid on insulin signaling 10, 23. Interestingly, the expansion of adiposity caused by TZDs in humans occurs in the subcutaneous adipose tissue, not in the visceral depots 23. Triglycerides stored in the subcutaneous fat are less lipolytic and have less access to portal circulation and the liver 23. In addition, PPARγ activation in adipose tissue regulates the production of adiponectin, resistin, and tumor necrosis factor-α (TNF-α). These secreted adipokines can impact insulin sensitivity in skeletal muscle and liver 10, 23. Taken together, adipose tissue is considered as the primary target of TZD action. In obese, diabetic mice with liver-specific PPARγ deletion, TZDs remain effective in lowering glucose levels, indicating that liver PPARγ is not a target 52. Since skeletal muscle accounts for 80% of the insulin-stimulated glucose disposal in humans, skeletal muscle-specific PPARγ knockout mice were used to address whether this tissue is also targeted by TZDs. Two studies have come to different conclusions 53, 54; the reason for this discrepancy is currently unclear.
Pathological roles of PPARγ in non-adipose tissues
In contrast to adipose tissue, liver and heart express very little PPARγ. However, under certain pathological conditions, these tissues can express considerable amounts of PPARγ that have significant impacts on metabolic homeostasis and tissue function.
Hepatic PPARγ expression is markedly increased in several obese or diabetic mouse models that are associated with liver steatosis 23. Studies suggest the hepatic PPARγ has a similar lipid deposition function as in adipose tissue. Ectopic adenoviral expression of PPARγ in the liver of lean mice causes liver steatosis 55. In the obese (ob/ob) mice, when PPARγ is specifically deleted in the liver, hepatic triglyceride level is normalized and fatty liver phenotype is ameliorated, accompanied by increased plasma fasting triglyceride and free fatty acid levels 52. TZD treatment further exacerbates the development of fatty liver in ob/ob mice, but not in ob/ob mice with hepatic PPARγ deletion 52. These results suggest that the elevated expression of hepatic PPARγ may be responsible for liver steatosis in these mouse models. In contrast, small clinical studies show that TZD treatment reduces hepatic lipid level in humans with nonalcoholic fatty liver 56, 57, indicating that PPARγ function in the adipose tissue continues to be dominant. However, the hepatic levels of PPARγ in these human subjects are not known.
In the heart, fatty acid uptake and oxidation are tightly coordinated. Upregulation of PPARγ causes metabolic perturbations that link to cardiomyopathies, diseases that can ultimately lead to heart failure. In ventricular samples of humans and mice with hypertrophic cardiomyopathy, levels of hypoxia-inducible factor α (HIFα) and PPARγ are increased 58. Mouse studies suggest that under hypertrophic stress, HIFα induces heart PPARγ expression. The concerted actions of HIFα and PPARγ then lead to cardiac lipid accumulation, apoptosis, and heart dysfunction 58. Another study reported that human patients with metabolic syndrome and aortic stenosis express high levels of PPARγ in the heart, which is strongly correlated with cardiac lipid accumulation and poor cardiac function 59. To directly examine the consequence of increased PPARγ, Goldberg and co-works overexpressed PPARγ in the heart and found that these transgenic mice develop steatosis and dilated cardiomyopathy. Treatment of the transgenic mice with the TZD drug further worsens these phenotypes 60. These studies together suggest that heart PPARγ, when its level is high under certain pathological conditions, may cause lipid overload that contributes to common forms of cardiomyopathy. One side effect of the TZD drugs in treatment of human diabetic patients is heart failure. While this could be due to fluid retention 49, the involvement of heart PPARγ in subsets of patients should be considered as well.
PPARα regulates hepatic lipid catabolism and fasting response
PPARα is expressed in many metabolically active tissues, but is high in the liver 61. The effects of PPARα agonist in vivo are mainly manifested in the liver 62. The PPARα null mice display a fatty liver phenotype resulted from a decreased expression of fatty acid oxidation genes 63, 64, 65, 66, 67. Indeed, the major function of PPARα is to promote fatty acid utilization. PPARα upregulates many genes involved in important steps of this process; these include genes for hepatic clearance of very low-density lipoprotein, fatty acid uptake, fatty acid activation and transport into the mitochondria, peroxisomal and mitochondrial β-oxidation, and some enzymes for mitochondrial respiration 68, 69, 70. The fibrate drugs used for the treatment of dyslipidemia are agonists of PPARα. Through activation of lipid catabolism, fibrates consistently lower plasma triglycerides in patients. Fibrates also raise plasma HDL through induction of apolipoprotein apoA-I and apoA-II in humans 68, 69. In addition, PPARα upregulates genes for fatty acid and triglyceride synthesis in the liver 70; the functional importance of this regulation is unknown.
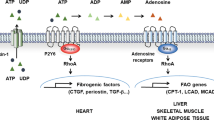
Ketogenesis in the liver is critical to the adaptive response to fasting. During fasting, fatty acids are mobilized in the adipose tissue and uptaken into the liver, where they are oxidized in the mitochondria to produce ketone bodies that provide the large energy source for other tissues. PPARα is induced by fasting and is required for ketogenesis 66. While the phenotype of the PPARα null mice at fed state is mild (fatty liver), fasting causes a very severe phenotype. This includes massive, exacerbated fatty liver, hypoketonemia, hypoglycemia, hypothermia, increased plasma free fatty acids, and loss of induction of fatty acid oxidation genes by fasting, suggesting that ketogenesis is significantly impaired 66, 67. Surprisingly, the hormonal peptide fibroblast growth factor 21 (FGF21) appears to be a key component downstream of PPARα. FGF21 is robustly induced during fasting in a large part in a PPARα-dependent manner 71, 72, 73. Introduction of FGF21 into PPARα null mice partially rescues hypoketonemia and fatty liver 73. On the other hand, knockdown of FGF21 in the liver causes fatty liver, increased plasma free fatty acids and decreased ketone bodies, when mice are fed a ketogenic diet 71. It was shown that FGF21 regulates the expression of lipases important for fatty acid release in the adipose tissue and the expression of a subset of PPARα target genes important for fatty acid oxidation in the liver 71, 73 (Figure 2).
An integrated model of PPARα-regulated ketogenesis. Fasting induces the expression of PPARα and PGC-1α. Activation of PPARα requires de novo synthesis of a PPARα ligand (16:0/18:1 GPC). Together, they induce the expression of fatty acid oxidation genes and FGF21. FGF21 in turn promotes lipolysis in the adipose tissue. The released free fatty acids (FFAs) are used as substrates for ketogenesis.
The PPARα-regulated ketogenesis probably requires PGC-1α co-activation. As a co-activator, PGC-1α is able to interact with and co-activate many transcription factors in vitro, including PPARs. Besides being a central regulator in brown fat metabolism as discussed earlier, PGC-1α also plays important roles in energy metabolism in the liver, skeletal muscle and heart 74. During periods of fasting, hepatic PGC-1α is induced, which in turn regulates fatty acid catabolism and gluconeogenesis 75. PGC-1α null mice exhibit fasting-induced fatty liver 76. A direct role of PGC-1α in fasting response was recently demonstrated by a study with liver-specific PGC-1α knockout mice 77. In this study, to mimic the physiological and pathological fluctuations of hepatic PGC-1α level, one allele of PGC-1α was deleted from the liver by Cre/Lox system; another allele remained intact. These PGC-1α liver heterozygous mice develop fasting-induced fatty liver, impaired ketogenesis, and have reduced expression of hepatic fatty acid oxidation genes, a phenotype that remarkably overlaps with PPARα null mice, indicating that PGC-1α-mediated activation of PPARα is likely to be a major mechanism for hepatic fatty acid catabolism (Figure 2). Transcriptional co-factors Lipin1, Sirt1 and BAF60α were shown to be involved as well 78, 79, 80.
A physiologically relevant endogenous PPARα ligand in the liver
Many endogenous lipid species have been considered as ligands for PPARs. However, they usually require presence in micromolar concentration range for PPAR activation, and most importantly, until recently, it has not been shown that lack of any of these lipids would affect the function of PPARs. The recent work by Chakravarthy et al. 81 identified the phospholipid 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine (16:0/18:1 GPC) as a physiologically relevant PPARα ligand in liver. The initial hint of the existence of a physiological ligand for PPARα came from their previous study on the liver-specific fatty acid synthase (FAS) knockout mice (FASKOL) 82. When these animals were subjected to fasting or placed on a diet that contained no fat (zero-fat diet), they developed a phenotype very similar to PPARα null mice, although the level of PPARα was not affected. Moreover, the phenotype was corrected when these mice were administered with a PPARα agonist. These results are consistent with the idea that FAS is required for the hepatic production of a PPARα ligand that can otherwise be derived from fat in normal diets, but not from fat released from peripheral tissues 82. To identify this ligand, the authors immunoprecipitated PPARα from liver and analyzed PPARα-associated lipids. The phospholipid 16:0/18:1 GPC was found to be associated with PPARα in the sample from wild-type mice, but not in that from FASKOL mice. This phospholipid potently binds to PPARα, weakly to PPARδ, and has no interaction with PPARγ in vitro. Knockdown of CEPT1, a gene required for the synthesis of 16:0/18:1 GPC, downregulates expression of some PPARα target genes in hepatic cells. Infusion of this lipid into mice reduces the liver triglyceride content in a PPARα-dependent manner. These results suggest that 16:0/18:1 GPC is an endogenous PPARα ligand that is indispensable for hepatic PPARα activation and function during fasting (Figure 2). It remains to be determined whether 16:0/18:1 GPC is also a physiological ligand for PPARα in other tissues. Moreover, structural analysis should provide insights into its differential binding affinity with the three PPARs.
PPARα in the skeletal muscle and heart: some unexpected phenotypes
In the PPARα knockout mice, muscle energy metabolism remains normal 83. A gain-of-function study was performed to address the potential role of PPARα in this tissue 84. Overexpression of PPARα in the skeletal muscle increases the expression of genes for fatty acid uptake and oxidation, but decreases the expression of the glucose transporter Glut4. Strikingly, under normal chow diet, these transgenic mice have elevated levels of plasma glucose and insulin, are glucose intolerant and insulin resistant; this occurs in the context of normal muscular lipid content and normal insulin signaling. When fed a high-fat diet, the transgenic mice, although resistant to obesity, have significantly higher muscular triglyceride content than controls, and continue to be glucose intolerant and insulin resistant. The effect of the transgene on muscular lipid accumulation is quite unexpected, given the known function of PPARα in fatty acid utilization; a plausible explanation is that the increased fatty acid uptake exceeds the increased muscle capacity for oxidation. This study confirms the role of PPARα in fatty acid catabolism in peripheral tissues. However, it is unclear what causes the severely impaired glucose homeostasis under normal diets. Is it due to the reduced expression of Glut4? Notably, skeletal muscle-specific deletion of Glut4 produces a similar defect in glucose homeostasis 85. It will be interesting to see whether the PPARα agonists also decrease Glut4 expression or promote lipid accumulation in the skeletal muscle. This question should be readily assessable with the PPARα transgenic mice because of the high expression of the transgene.
PPARα also regulates cardiac metabolism. PPARα null mice have decreased expression of cardiac fatty acid oxidation genes and decreased rates of fatty acid oxidation, but maintain normal cardiac function 83, 86. Similar to the skeletal muscle PPARα transgene, overexpression of PPARα in the heart increases the expression of genes for fatty acid transport and oxidation, and decreases Glut4 expression; this leads to lipid accumulation, reduced glucose transport and utilization, insulin resistance, and cardiomyopathy 87, 88. These phenotypes are exacerbated on high-fat diets 89, and are corrected by deficiency of the fatty acid transporter CD36 without affecting fatty acid oxidation 90. Interestingly, Glut4 expression and glucose transport are increased in the PPARγ cardiac transgenic mice, which also display lipid accumulation and cardiomyopathy 60. Moreover, in the PPARα muscle transgenic mice, decreased Glut4 expression and defects in glucose metabolism occur in the absence of lipid accumulation 84. These observations indicate that the decreased Glut4 expression and glucose utilization in the PPARα transgenic mice is a PPAR-isoform-specific phenotype not caused by lipid accumulation or increased fatty acid oxidation per se. If this is true, then how this phenotype is corrected by CD36 deficiency is unknown.
PPARδ is an integral component in a transcriptional network regulating brown fat metabolism
PPARδ is ubiquitously expressed in many tissues. Genetic and pharmacological studies reveal its role as a powerful regulator of fatty acid catabolism and energy homeostasis 91. Initially, due to the unavailability of a specific PPARδ agonist, we used transgenic mice expressing an activated form of PPARδ by fusion with a VP16 activation domain to uncover its tissue-specific function 92. Adipose tissue expression of this transgene produces lean mice that are resistant to obesity and tissue steatosis induced either genetically or by a high-fat diet. Although WAT expressed a slightly higher level of the transgene than BAT, the induction of the expression of fat-burning genes mainly occurs in BAT. Interestingly, UCP1 induction is the highest among all the genes examined. In WAT, only UCP1 is induced. The reason for this selective induction in BAT vs WAT is unknown. This work helped demonstrate a potential in vivo function of brown fat PPARδ in energy expenditure.
To further study the role of PPARδ in brown fat metabolism, we generated an immortalized brown fat preadipocyte cell line from mice containing floxed PPARδ alleles. In vitro deletion of the PPARδ alleles by cre recombinase adenovirus-mediated excision, as expected, does not affect brown fat cell differentiation, yet significantly downregulates the expression of fat-burning genes 93. In particular, UCP1 expression is the mostly decreased, typically by more than 10-fold, while the levels of PGC-1α, PPARγ and PPARα remain unchanged. This led us to test whether PPARδ acts downstream of PGC-1α. The β3-adrenergic receptor agonist stimulates UCP1 expression through PGC-1α. The PPARδ-deficient brown fat cells are almost completely unresponsive to the β3-adrenergic receptor agonist in the induction of UCP1. Moreover, in the absence of PPARδ, PGC-1α is longer associated with the PPAR-binding site of the UCP1 promoter. Finally, the ability of the PPARδ-fat knockout mice to maintain body temperature during cold exposure is compromised. These results suggest that, in the brown fat, there is a cell-autonomous requirement for PPARδ in the expression of at least some of the PGC-1α target genes and that PPARδ at least partly mediates the action of PGC-1α.
The PGC-1α/PPARδ-regulated brown fat metabolism is fine-tuned by twist-1 93. Twist-1 is a critical, physiological negative regulator in brown fat metabolism. It is selectively expressed in the adipose tissue, interacts with PGC-1α, and is recruited to the promoters of PGC-1α's target genes to suppress PGC-1α function. Indeed, twist-1 heterozygous knockout mice are resistant to high fat-induced obesity, whereas the fat twist-1 transgenic mice have the opposite phenotype. Interestingly, upon activation by its ligand, PPARδ is recruited to the twist-1 promoter and induces twsit-1 expression both in vitro and in vivo, suggesting a negative-feedback mechanism. Thus, PPARδ is an integral component in brown fat metabolism by coordinating the actions of PGC-1α and twist-1.
PPARδ in the skeletal muscle and heart: a comparison with PPARα
Muscle fibers are broadly classified into two groups, oxidative fibers and glycolytic fibers. Oxidative fibers have high mitochondrial content and mainly use mitochondrial oxidative metabolism for energy production, whereas glycolytic fibers have low mitochondrial content and rely on glycolytic metabolism as a major energy source. PPARδ has a higher expression than PPARα in the skeletal muscle 61, 83 and is more abundant in the oxidative fibers 94, 95. Treatment of skeletal muscle cells with the PPARδ agonists in vitro induces the expression of genes for fatty acid catabolism and promotes fatty acid oxidation 83, 92, 96. To test the in vivo function of PPARδ, skeletal muscle PPARδ transgenic mice expressing either a wild type or activated form of PPARδ were generated 94, 97. There is a remarkable increase of oxidative fibers in these transgenic mice, similar to that observed in the skeletal muscle PGC-1α transgenic mice 98; however, PGC-1α is not induced in the PPARδ transgenic mice. While the phenotype of the transgenic mice expressing the wild-type PPARδ remains to be fully characterized, the transgenic mice expressing the activated PPARδ are found to be completely protected against high-fat diet-induced obesity, muscular lipid accumulation, hyperinsulinemia, glucose intolerance, and insulin resistance 94 (our unpublished data). The normalization of insulin sensitivity by the PPARδ transgene on the high-fat diet is most likely due to the lack of muscular lipid accumulation. While both the PPARδ and PPARα muscle transgenic lines are resistant to high-fat diet-induced obesity, the PPARα line, in striking contrast, has increased muscular lipid accumulation and is insulin resistant and glucose intolerant (Figure 3). Both of these two lines display upregulation of genes for fatty acid uptake, β-oxidation, and mitochondrial respiration 84, 94, 97 (our unpublished data). Moreover, although the PPARδ transgene is constitutively active, its expression level is very low; instead, endogenous muscle PPARδ expression is highly elevated so that it may at least partly contribute to the phenotype 94. Thus, what causes the phenotypic differences between the PPARδ and PPARα transgenic mice is currently unclear and can be either isoform-specific target gene expression or their overall differential effects on metabolic programs. Interestingly, the skeletal muscle PGC-1α transgenic mice, although showing increased oxidative fibers, not only are not protected against high-fat diet-induced body weight gain, but also are more insulin resistant due to muscular lipid accumulation, compared to control mice 98, 99.
A phenotype opposite to that of PPARδ transgenic mice was revealed in the skeletal muscle-specific PPARδ knockout mice 100. Deletion of PPARδ decreases the expression of many genes for fatty acid β-oxidation and mitochondrial respiration function, and increases the number of muscle fibers with lower oxidative capacity, leading to an increased body weight gain and insulin resistance. The skeletal muscle of these knockouts also shows a 50% decrease of PGC-1α that could be responsible for the muscle phenotype. However, it is also possible that the decrease in PGC-1α is a secondary chronic effect caused by the increased number of less oxidative fibers. As a similar muscle phenotype was observed when PPARδ was deleted by an inducible cre after the muscle was formed 100, it will be interesting to examine the level of PGC-1α in this setting. Nevertheless, this work demonstrates that PPARδ is required for the formation and maintenance of oxidative muscle fibers.
Cardiac-specific deletion of PPARδ causes lipid accumulation and cardiomyopathy 101. Unlike PPARα and PPARγ, overexpression of PPARδ in the heart is not associated with lipid accumulation and has no detrimental effects on cardiac function 102. Surprisingly, the transgene increases Glut4 expression and promotes glucose transport in the heart 102 (Figure 4).
Function and mechanism of PPARδ agonist in fat burning
An important question is whether activation of endogenous PPARδ by its agonist would be metabolically beneficial. Treatment of insulin-resistant, obese monkeys for four weeks with the PPARδ agonist lowers plasma insulin and triglyceride levels and increases HDL level, but has no effect on body weight 103. Increased fatty acid oxidation was observed in the skeletal muscle and BAT, but not in the liver, of PPARδ agonist-treated mice 104. The PPARδ agonist effectively prevents the high-fat diet-induced obesity, hyperinsulinemia, tissue steatosis, glucose intolerance, and insulin resistance 94, 104. When given to the diabetic, obese ob/ob mice, the PPARδ agonist produces similar beneficial effects, along with a significant decrease in blood glucose level 104, 105. However, the body weight reduction in the ob/ob mice is very modest 104, indicating that the agonist is less effective on predisposed obesity. Overall, the PPARδ agonist elicits profound metabolic benefits that essentially recapitulate the phenotypes of the transgenic mice (Figure 3). Future studies with conditional PPARδ knockout mice are needed to address the relative contribution of different tissues to the agonist effects. Presumably, skeletal muscle and BAT are major target tissues.
Is the PPARδ agonist useful for human metabolic syndrome? To date, this has not been extensively studied or reported. In a small pilot study, PPARδ agonist was given to healthy moderately obese subjects for two weeks. Administration of the PPARδ agonist significantly reduces fasting plasma triglycerides, LDL cholesterol, insulin, and liver fat content; in contrast, these changes were not observed in subjects when given the PPARα agonist, except for the lowering of plasma triglycerides 106. Whether these beneficial effects of the PPARδ agonist can also be observed in a large study remains to be seen.
Conceivably, fat burning in the skeletal muscle requires coordination between fatty acid β-oxidation and mitochondrial respiration. It is clear that PPARδ agonists enhance fatty acid β-oxidation. Are they also capable of promoting mitochondrial respiration function and thereby driving the formation of oxidative muscle fibers? Treatment of fibroblasts or myoblasts in vitro with the PPARδ agonist increases the activities of enzymes in the mitochondrial respiration chain 107. In a mouse model of mitochondrial myopathy, bezafibrate, an agonist for both PPARδ and PPARα, elevates the levels of proteins for mitochondrial respiration, increases mitochondrial biogenesis, and improves the myopathy phenotype 108. Treatment of wild-type mice with the PPARδ agonist promotes the formation of oxidative muscle fibers based on succinate dehydrogenase activity staining 109. However, other in vivo studies did not observe any effect of the agonist on expression of a group of genes for mitochondrial respiration function 105 or on the formation of oxidative fibers based on ATPase activity staining 110; but combined with exercise training, the agonist increases the number of oxidative fibers 110. Thus, it is likely that PPARδ and its agonist may only regulate a subset of genes for mitochondrial respiration function and that a systematic effect requires additional signaling inputs. Notably, levels of PPARδ and PGC-1α in the skeletal muscle are elevated in several genetic mouse models that display increased mitochondrial biogenesis and oxidative fibers 111, 112.
PPARδ agonists activate gene expression through facilitating the recruitment of transcriptional co-activators. In primary, PGC-1α-deficient myotubes, PPARδ agonist-induced fatty acid oxidation is attenuated but not abolished. These data indicate that co-activation of PPARδ by PGC-1α is partly responsible for the ligand effects in the skeletal muscle and that a PGC-1α-independent mechanism also exits 105. In agreement with this, we have found that in C2C12 muscle cells where there is little expression of PGC-1α, PPARδ agonist-induced gene expression and fatty acid oxidation are not suppressed by ectopic expression of the PGC-1α inhibitor, twist-1 (our unpublished data).
PPARδ and exercise physiology
Muscle fibers are strongly associated with exercise physiology 113. On one hand, oxidative muscle fibers confer exercise endurance; on the other hand, regular exercise training promotes the formation of oxidative fibers, which leads to the improvement in metabolic syndrome. Given the effects of PPARδ on muscle fiber and energy metabolism, two related questions become apparent: whether activation of PPARδ promotes exercise endurance and whether PPARδ mediates some of the metabolic benefits of exercise training. The first question has been addressed. When placed on a treadmill, muscle PPARδ transgenic mice are resistant to fatigue and are able to run twice the distance of wild-type mice 94. Conversely, the muscle PPARδ knockout mice run about 30% less the distance of the wild-type mice 100. The PPARδ agonist itself is not sufficient to promote exercise performance; however, when combined with exercise training, the agonist significantly increases endurance compared to exercise alone 110. Part of the molecular mechanism underlying this synergistic effect on endurance is that exercise activates the AMPK kinase, which further enhances the ligand-activated PPARδ transcriptional activity 110. This work reveals the AMPK-PPARδ axis as a pharmacological target to reprogram muscle endurance. Does PPARδ in turn contribute to the metabolic benefits pertinent to exercise? This has not been addressed genetically. However, PPARδ expression in the skeletal muscle is increased by exercise training in both rodents and humans 97, 114, 115. Importantly, there is a strong correlation between exercise-induced PPARδ expression and improvement of clinical parameters in type 2 diabetic patients 115. These observations, along with the known effects of PPARδ activation on energy metabolism, suggest a formal possibility that PPARδ may mediate some of the metabolic benefits of exercise.
Conclusions
The three PPARs, by acting as lipid sensors, are major metabolic regulators in the body and together they control almost every aspect of fatty acid metabolism (Figure 5). Importantly, through regulation of lipid metabolism, these receptors greatly impact insulin sensitivity and glucose homeostasis. Functional impairment or dysregulation of these receptors is shown to lead to obesity, lipodystrophy, fatty liver, insulin resistance, type 2 diabetes, or cardiomyopathy, whereas proper activation by ligands offers metabolic benefits. These studies have helped significantly in understanding the transcriptional regulation of energy metabolism and the pathogenesis of metabolic syndrome. However, many interesting questions remain. For example, what are the detailed molecular mechanisms employed by these receptors in gene regulation? What are the causes for the side effects of the PPAR drugs? What are the endogenous, physiological ligands in different tissues? How do the PPARs and their potential endogenous ligands respond to environmental signals? How are their actions coordinated? The three PPARs share a large overlap of target gene profile, but the ensuing phenotypes can be very different. This is probably best exemplified in the skeletal muscle and heart of transgenic mice. How are their distinct functions specified? Addressing these questions will be important for us to fully understand the roles of these receptors in energy metabolism and metabolic syndrome.
References
Reaven GM . The insulin resistance syndrome: definition and dietary approaches to treatment. Annu Rev Nutr 2005; 25:391–406.
Stumvoll M, Goldstein BJ, van Haeften TW . Type 2 diabetes: principles of pathogenesis and therapy. Lancet 2005; 365:1333–1346.
Savage DB, Petersen KF, Shulman GI . Disordered lipid metabolism and the pathogenesis of insulin resistance. Physiol Rev 2007; 87:507–520.
Schenk S, Saberi M, Olefsky JM . Insulin sensitivity: modulation by nutrients and inflammation. J Clin Invest 2008; 118:2992–3002.
Wellen KE, Hotamisligil GS . Inflammation, stress, and diabetes. J Clin Invest 2005; 115:1111–1119.
Bensinger SJ, Tontonoz P . Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature 2008; 454:470–477.
Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ . Nuclear receptors and lipid physiology: opening the X-files. Science 2001; 294:1866–1870.
Odegaard JI, Ricardo-Gonzalez RR, Red Eagle A, et al. Alternative M2 activation of Kupffer cells by PPARdelta ameliorates obesity-induced insulin resistance. Cell Metab 2008; 7:496–507.
Kang K, Reilly SM, Karabacak V, et al. Adipocyte-derived Th2 cytokines and myeloid PPARdelta regulate macrophage polarization and insulin sensitivity. Cell Metab 2008; 7:485–495.
Anghel SI, Wahli W . Fat poetry: a kingdom for PPAR gamma. Cell Res 2007; 17:486–511.
Rosen ED, Spiegelman BM . Adipocytes as regulators of energy balance and glucose homeostasis. Nature 2006; 444:847–853.
Farmer SR . Transcriptional control of adipocyte formation. Cell Metab 2006; 4:263–273.
Rosen ED, MacDougald OA . Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 2006; 7:885–896.
Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM . mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev 1994; 8:1224–1234.
Tontonoz P, Hu E, Spiegelman BM . Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 1994; 79:1147–1156.
Rosen ED, Hsu CH, Wang X, et al. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev 2002; 16:22–26.
Rosen ED, Sarraf P, Troy AE, et al. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell 1999; 4:611–617.
Barak Y, Nelson MC, Ong ES, et al. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell 1999; 4:585–595.
Koutnikova H, Cock TA, Watanabe M, et al. Compensation by the muscle limits the metabolic consequences of lipodystrophy in PPAR gamma hypomorphic mice. Proc Natl Acad Sci USA 2003; 100:14457–14462.
Imai T, Takakuwa R, Marchand S, et al. Peroxisome proliferator-activated receptor gamma is required in mature white and brown adipocytes for their survival in the mouse. Proc Natl Acad Sci USA 2004; 101:4543–4547.
Jones JR, Barrick C, Kim KA, et al. Deletion of PPARgamma in adipose tissues of mice protects against high fat diet-induced obesity and insulin resistance. Proc Natl Acad Sci USA 2005; 102:6207–6212.
He W, Barak Y, Hevener A, et al. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci USA 2003; 100:15712–15717.
Semple RK, Chatterjee VK, O'Rahilly S . PPAR gamma and human metabolic disease. J Clin Invest 2006; 116:581–589.
Tanaka T, Yoshida N, Kishimoto T, Akira S . Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J 1997; 16:7432–7443.
Linhart HG, Ishimura-Oka K, DeMayo F, et al. C/EBPalpha is required for differentiation of white, but not brown, adipose tissue. Proc Natl Acad Sci USA 2001; 98:12532–12537.
Wu Z, Rosen ED, Brun R, et al. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell 1999; 3:151–158.
El-Jack AK, Hamm JK, Pilch PF, Farmer SR . Reconstitution of insulin-sensitive glucose transport in fibroblasts requires expression of both PPARgamma and C/EBPalpha. J Biol Chem 1999; 274:7946–7951.
Lefterova MI, Zhang Y, Steger DJ, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev 2008; 22:2941–2952.
Cho YW, Hong S, Jin Q, et al. Histone methylation regulator PTIP is required for PPARgamma and C/EBPalpha expression and adipogenesis. Cell Metab 2009; 10:27–39.
Wang W, Huang L, Huang Y, et al. Mediator MED23 links insulin signaling to the adipogenesis transcription cascade. Dev Cell 2009; 16:764–771.
Chen Z, Torrens JI, Anand A, Spiegelman BM, Friedman JM . Krox20 stimulates adipogenesis via C/EBPbeta-dependent and -independent mechanisms. Cell Metab 2005; 1:93–106.
Oishi Y, Manabe I, Tobe K, et al. Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab 2005; 1:27–39.
Sha H, He Y, Chen H, et al. The IRE1alpha-XBP1 pathway of the unfolded protein response is required for adipogenesis. Cell Metab 2009; 9:556–564.
Lowell BB, Spiegelman BM . Towards a molecular understanding of adaptive thermogenesis. Nature 2000; 404:652–660.
Cannon B, Nedergaard J . Brown adipose tissue: function and physiological significance. Physiol Rev 2004; 84:277–359.
van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med 2009; 360:1500–1508.
Cypess AM, Lehman S, Williams G, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med 2009; 360:1509–1517.
Virtanen KA, Lidell ME, Orava J, et al. Functional brown adipose tissue in healthy adults. N Engl J Med 2009; 360:1518–1525.
Seale P, Bjork B, Yang W, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature 2008; 454:961–967.
Seale P, Kajimura S, Yang W, et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab 2007; 6:38–54.
Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM . A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 1998; 92:829–839.
Kajimura S, Seale P, Kubota K, et al. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature 2009; 460:1154–1158.
Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM . Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab 2006; 3:333–341.
Scime A, Grenier G, Huh MS, et al. Rb and p107 regulate preadipocyte differentiation into white versus brown fat through repression of PGC-1alpha. Cell Metab 2005; 2:283–295.
Powelka AM, Seth A, Virbasius JV, et al. Suppression of oxidative metabolism and mitochondrial biogenesis by the transcriptional corepressor RIP140 in mouse adipocytes. J Clin Invest 2006; 116:125–136.
Christian M, Kiskinis E, Debevec D, Leonardsson G, White R, Parker MG . RIP140-targeted repression of gene expression in adipocytes. Mol Cell Biol 2005; 25:9383–9391.
Tseng YH, Kokkotou E, Schulz TJ, et al. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature 2008; 454:1000–1004.
Olefsky JM . Treatment of insulin resistance with peroxisome proliferator-activated receptor gamma agonists. J Clin Invest 2000; 106:467–472.
Mudaliar S, Chang AR, Henry RR . Thiazolidinediones, peripheral edema, and type 2 diabetes: incidence, pathophysiology, and clinical implications. Endocr Pract 2003; 9:406–416.
Lehmann JM, Moore LB, Smith-Oliver TA, Wilkison WO, Willson TM, Kliewer SA . An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma). J Biol Chem 1995; 270:12953–12956.
Harris PK, Kletzien RF . Localization of a pioglitazone response element in the adipocyte fatty acid-binding protein gene. Mol Pharmacol 1994; 45:439–445.
Matsusue K, Haluzik M, Lambert G, et al. Liver-specific disruption of PPARgamma in leptin-deficient mice improves fatty liver but aggravates diabetic phenotypes. J Clin Invest 2003; 111:737–747.
Hevener AL, He W, Barak Y, et al. Muscle-specific Pparg deletion causes insulin resistance. Nat Med 2003; 9:1491–1497.
Norris AW, Chen L, Fisher SJ, et al. Muscle-specific PPARgamma-deficient mice develop increased adiposity and insulin resistance but respond to thiazolidinediones. J Clin Invest 2003; 112:608–618.
Yu S, Matsusue K, Kashireddy P, et al. Adipocyte-specific gene expression and adipogenic steatosis in the mouse liver due to peroxisome proliferator-activated receptor gamma1 (PPARgamma1) overexpression. J Biol Chem 2003; 278:498–505.
Belfort R, Harrison SA, Brown K, et al. A placebo-controlled trial of pioglitazone in subjects with nonalcoholic steatohepatitis. N Engl J Med 2006; 355:2297–2307.
Promrat K, Lutchman G, Uwaifo GI, et al. A pilot study of pioglitazone treatment for nonalcoholic steatohepatitis. Hepatology 2004; 39:188–196.
Krishnan J, Suter M, Windak R, et al. Activation of a HIF1alpha-PPARgamma axis underlies the integration of glycolytic and lipid anabolic pathways in pathologic cardiac hypertrophy. Cell Metab 2009; 9:512–524.
Marfella R, Di Filippo C, Portoghese M, et al. Myocardial lipid accumulation in patients with pressure-overloaded heart and metabolic syndrome. J Lipid Res 2009; 50:2314–2323.
Son NH, Park TS, Yamashita H, et al. Cardiomyocyte expression of PPARgamma leads to cardiac dysfunction in mice. J Clin Invest 2007; 117:2791–2801.
Braissant O, Foufelle F, Scotto C, Dauca M, Wahli W . Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology 1996; 137:354–366.
Cook WS, Yeldandi AV, Rao MS, Hashimoto T, Reddy JK . Less extrahepatic induction of fatty acid beta-oxidation enzymes by PPAR alpha. Biochem Biophys Res Commun 2000; 278:250–257.
Aoyama T, Peters JM, Iritani N, et al. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor alpha (PPARalpha). J Biol Chem 1998; 273:5678–5684.
Akiyama TE, Nicol CJ, Fievet C, et al. Peroxisome proliferator-activated receptor-alpha regulates lipid homeostasis, but is not associated with obesity: studies with congenic mouse lines. J Biol Chem 2001; 276:39088–39093.
Sugden MC, Bulmer K, Gibbons GF, Knight BL, Holness MJ . Peroxisome-proliferator-activated receptor-alpha (PPARalpha) deficiency leads to dysregulation of hepatic lipid and carbohydrate metabolism by fatty acids and insulin. Biochem J 2002; 364:361–368.
Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W . Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest 1999; 103:1489–1498.
Leone TC, Weinheimer CJ, Kelly DP . A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci USA 1999; 96:7473–7478.
Lefebvre P, Chinetti G, Fruchart JC, Staels B . Sorting out the roles of PPAR alpha in energy metabolism and vascular homeostasis. J Clin Invest 2006; 116:571–580.
Duval C, Muller M, Kersten S . PPARalpha and dyslipidemia. Biochim Biophys Acta 2007; 1771:961–971.
Rakhshandehroo M, Hooiveld G, Muller M, Kersten S . Comparative analysis of gene regulation by the transcription factor PPARalpha between mouse and human. PLoS One 2009; 4:e6796.
Badman MK, Pissios P, Kennedy AR, Koukos G, Flier JS, Maratos-Flier E . Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab 2007; 5:426–437.
Lundasen T, Hunt MC, Nilsson LM, et al. PPARalpha is a key regulator of hepatic FGF21. Biochem Biophys Res Commun 2007; 360:437–440.
Inagaki T, Dutchak P, Zhao G, et al. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab 2007; 5:415–425.
Lin J, Handschin C, Spiegelman BM . Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab 2005; 1:361–370.
Yoon JC, Puigserver P, Chen G, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 2001; 413:131–138.
Leone TC, Lehman JJ, Finck BN, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol 2005; 3:e101.
Estall JL, Kahn M, Cooper MP, et al. Sensitivity of lipid metabolism and insulin signaling to genetic alterations in hepatic peroxisome proliferator-activated receptor-gamma coactivator-1alpha expression. Diabetes 2009; 58:1499–1508.
Li S, Liu C, Li N, et al. Genome-wide coactivation analysis of PGC-1alpha identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metab 2008; 8:105–117.
Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X . Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab 2009; 9:327–338.
Finck BN, Gropler MC, Chen Z, et al. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab 2006; 4:199–210.
Chakravarthy MV, Lodhi IJ, Yin L, et al. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell 2009; 138:476–488.
Chakravarthy MV, Pan Z, Zhu Y, et al. “New” hepatic fat activates PPARalpha to maintain glucose, lipid, and cholesterol homeostasis. Cell Metab 2005; 1:309–322.
Muoio DM, MacLean PS, Lang DB, et al. Fatty acid homeostasis and induction of lipid regulatory genes in skeletal muscles of peroxisome proliferator-activated receptor (PPAR) alpha knock-out mice. Evidence for compensatory regulation by PPAR delta. J Biol Chem 2002; 277:26089–26097.
Finck BN, Bernal-Mizrachi C, Han DH, et al. A potential link between muscle peroxisome proliferator- activated receptor-alpha signaling and obesity-related diabetes. Cell Metab 2005; 1:133–144.
Zisman A, Peroni OD, Abel ED, et al. Targeted disruption of the glucose transporter 4 selectively in muscle causes insulin resistance and glucose intolerance. Nat Med 2000; 6:924–928.
Campbell FM, Kozak R, Wagner A, et al. A role for peroxisome proliferator-activated receptor alpha (PPARalpha) in the control of cardiac malonyl-CoA levels: reduced fatty acid oxidation rates and increased glucose oxidation rates in the hearts of mice lacking PPARalpha are associated with higher concentrations of malonyl-CoA and reduced expression of malonyl-CoA decarboxylase. J Biol Chem 2002; 277:4098–4103.
Finck BN, Lehman JJ, Leone TC, et al. The cardiac phenotype induced by PPARalpha overexpression mimics that caused by diabetes mellitus. J Clin Invest 2002; 109:121–130.
Park SY, Cho YR, Finck BN, et al. Cardiac-specific overexpression of peroxisome proliferator-activated receptor-alpha causes insulin resistance in heart and liver. Diabetes 2005; 54:2514–2524.
Finck BN, Han X, Courtois M, et al. A critical role for PPARalpha-mediated lipotoxicity in the pathogenesis of diabetic cardiomyopathy: modulation by dietary fat content. Proc Natl Acad Sci USA 2003; 100:1226–1231.
Yang J, Sambandam N, Han X, et al. CD36 deficiency rescues lipotoxic cardiomyopathy. Circ Res 2007; 100:1208–1217.
Barish GD, Narkar VA, Evans RM . PPARdelta: a dagger in the heart of the metabolic syndrome. J Clin Invest 2006; 116:590–597.
Wang YX, Lee CH, Tiep S, et al. Peroxisome-proliferator-activated receptor delta activates fat metabolism to prevent obesity. Cell 2003; 113:159–170.
Pan D, Fujimoto M, Lopes A, Wang YX . Twist-1 is a PPARdelta-inducible, negative-feedback regulator of PGC-1alpha in brown fat metabolism. Cell 2009; 137:73–86.
Wang YX, Zhang CL, Yu RT, et al. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol 2004; 2:e294.
Lunde IG, Ekmark M, Rana ZA, Buonanno A, Gundersen K . PPARdelta expression is influenced by muscle activity and induces slow muscle properties in adult rat muscles after somatic gene transfer. J Physiol 2007; 582:1277–1287.
Holst D, Luquet S, Nogueira V, Kristiansen K, Leverve X, Grimaldi PA . Nutritional regulation and role of peroxisome proliferator-activated receptor delta in fatty acid catabolism in skeletal muscle. Biochim Biophys Acta 2003; 1633:43–50.
Luquet S, Lopez-Soriano J, Holst D, et al. Peroxisome proliferator-activated receptor delta controls muscle development and oxidative capability. Faseb J 2003; 17:2299–2301.
Lin J, Wu H, Tarr PT, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature 2002; 418:797–801.
Choi CS, Befroy DE, Codella R, et al. Paradoxical effects of increased expression of PGC-1alpha on muscle mitochondrial function and insulin-stimulated muscle glucose metabolism. Proc Natl Acad Sci USA 2008; 105:19926–19931.
Schuler M, Ali F, Chambon C, et al. PGC1alpha expression is controlled in skeletal muscles by PPARbeta, whose ablation results in fiber-type switching, obesity, and type 2 diabetes. Cell Metab 2006; 4:407–414.
Cheng L, Ding G, Qin Q, et al. Cardiomyocyte-restricted peroxisome proliferator-activated receptor-delta deletion perturbs myocardial fatty acid oxidation and leads to cardiomyopathy. Nat Med 2004; 10:1245–1250.
Burkart EM, Sambandam N, Han X, et al. Nuclear receptors PPARbeta/delta and PPARalpha direct distinct metabolic regulatory programs in the mouse heart. J Clin Invest 2007; 117:3930–3939.
Oliver WR, Jr, Shenk JL, Snaith MR, et al. A selective peroxisome proliferator-activated receptor delta agonist promotes reverse cholesterol transport. Proc Natl Acad Sci USA 2001; 98:5306–5311.
Tanaka T, Yamamoto J, Iwasaki S, et al. Activation of peroxisome proliferator-activated receptor delta induces fatty acid beta-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc Natl Acad Sci USA 2003; 100:15924–15929.
Kleiner S, Nguyen-Tran V, Bare O, Huang X, Spiegelman B, Wu Z . PPARδ agonism activates fatty acid oxidation via PGC-1α but does not increase mitochondrial gene expression and function. J Biol Chem 2009; 284:18624–18633.
Riserus U, Sprecher D, Johnson T, et al. Activation of peroxisome proliferator-activated receptor (PPAR)delta promotes reversal of multiple metabolic abnormalities, reduces oxidative stress, and increases fatty acid oxidation in moderately obese men. Diabetes 2008; 57:332–339.
Bastin J, Aubey F, Rotig A, Munnich A, Djouadi F . Activation of peroxisome proliferator-activated receptor pathway stimulates the mitochondrial respiratory chain and can correct deficiencies in patients' cells lacking its components. J Clin Endocrinol Metab 2008; 93:1433–1441.
Wenz T, Diaz F, Spiegelman BM, Moraes CT . Activation of the PPAR/PGC-1alpha pathway prevents a bioenergetic deficit and effectively improves a mitochondrial myopathy phenotype. Cell Metab 2008; 8:249–256.
Gaudel C, Schwartz C, Giordano C, Abumrad NA, Grimaldi PA . Pharmacological activation of PPARbeta promotes rapid and calcineurin-dependent fiber remodeling and angiogenesis in mouse skeletal muscle. Am J Physiol Endocrinol Metab 2008; 295:E297–E304.
Narkar VA, Downes M, Yu RT, et al. AMPK and PPARdelta agonists are exercise mimetics. Cell 2008; 134:405–415.
Casas F, Pessemesse L, Grandemange S, et al. Overexpression of the mitochondrial T3 receptor p43 induces a shift in skeletal muscle fiber types. PLoS One 2008; 3:e2501.
Miyashita K, Itoh H, Tsujimoto H, et al. Natriuretic peptides/cGMP/cGMP-dependent protein kinase cascades promote muscle mitochondrial biogenesis and prevent obesity. Diabetes 2009; 58:2880–2892
Zierath JR, Hawley JA . Skeletal muscle fiber type: influence on contractile and metabolic properties. PLoS Biol 2004; 2:e348.
Mahoney DJ, Parise G, Melov S, Safdar A, Tarnopolsky MA . Analysis of global mRNA expression in human skeletal muscle during recovery from endurance exercise. FASEB J 2005; 19:1498–1500.
Fritz T, Kramer DK, Karlsson HK, et al. Low-intensity exercise increases skeletal muscle protein expression of PPARdelta and UCP3 in type 2 diabetic patients. Diabetes Metab Res Rev 2006; 22:492–498.
Acknowledgements
I am grateful to Dr Dongning Pan (University of Massachusetts Medical School) for the help on figure illustrations. This work was funded by grants from American Diabetes Association, Smith Family Foundation, and NIH/NIDDK (R01DK076118).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, YX. PPARs: diverse regulators in energy metabolism and metabolic diseases. Cell Res 20, 124–137 (2010). https://doi.org/10.1038/cr.2010.13
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2010.13
Keywords
This article is cited by
-
Harnessing peroxisome proliferator-activated receptor γ agonists to induce Heme Oxygenase-1: a promising approach for pulmonary inflammatory disorders
Cell Communication and Signaling (2024)
-
Exploring binding mode assessment of novel kaempferol, resveratrol, and quercetin derivatives with PPAR-α as potent drug candidates against cancer
Molecular Diversity (2023)
-
Targeting lipid-sensing nuclear receptors PPAR (α, γ, β/δ): HTVS and molecular docking/dynamics analysis of pharmacological ligands as potential pan-PPAR agonists
Molecular Diversity (2023)
-
Swimming exercise activates peroxisome proliferator-activated receptor-alpha and mitigates age-related renal fibrosis in rats
Molecular and Cellular Biochemistry (2023)
-
Endogenous renal adiponectin drives gluconeogenesis through enhancing pyruvate and fatty acid utilization
Nature Communications (2023)