Abstract
We used the promoter trap technique to identify a rice plant, named 107#, in which the β-glucuronidase (GUS) reporter gene was expressed specifically in the endosperm. A single copy of the T-DNA was inserted into the plant genome, and a candidate gene OsRRM was identified by the insertion. The OsRRM promoter directed GUS expression specifically in rice endosperm, analogous to the GUS expression pattern observed in 107#. OsRRM is a single-copy gene in rice and encodes a nuclear protein containing 1 005 amino-acid residues with two RNA recognition motifs and one Spen paralog and ortholog C-terminal domain. Western blot analysis confirmed that the OsRRM protein was specifically expressed in rice endosperm. Ectopic expression of OsRRM in transgenic plants led to abnormalities, such as short stature, retarded growth and low fructification rates. Our data, in conjunction with the reported function of Spen genes, implicated OsRRM in the regulation of cell development in rice endosperm.
Similar content being viewed by others
Introduction
Rice, a major source of carbohydrates of human food, produces diplontic embryo and triploid endosperm through double fertilization. Starch, which accounts for more than 90% of the dry matter in rice seed, is deposited in the endosperm. Endosperm development comprises several stages before it reaches maturation, including the syncytial, cellular, reserve-storage and programmed cell death stages. During this process, various plant hormones and environmental factors play important roles in regulating the expression of numerous genes. Research on endosperm development is important, given that the mechanisms of plant cell differentiation and apoptosis and reserve material accumulation in rice have not been fully elucidated 1.
Various techniques have been utilized to isolate and characterize essential genes in rice so as to better understand the molecular mechanisms regulating rice development. Large-scale mutant screening is widely used to identify mutants defective in different aspects of development. Chemicals, such as EMS 2, 3, and many physical treatments, including fast neutrons 4, have been used to generate random mutations in the plant genome. With the breakthrough of transformation technology in rice, transposon 5 and T-DNA tagging 6 have become powerful tools to isolate rice mutants and clone the corresponding genes.
Gene trap is an effective method to identify genes without the need for morphological phenotype alterations. By randomly introducing a reporter gene that lacks its own promoter into the plant genome, we can “trap” or tag an endogenous gene. The reporter gene will only be expressed if, by chance, it is inserted in the immediate vicinity of an endogenous gene promoter. The expression pattern of the “trapped” endogenous gene can then be predicted with the help of the reporter gene 7.
Spen (split ends) proteins are characterized by an N-terminal RNA recognition motif (RRM) domain and a structurally well-conserved C-terminal Spen paralog and ortholog C-terminal (SPOC) domain. While the molecular weight of Spen proteins varies greatly, ranging from 90 to 600 kDa, SPOC domain contains approximately 165 amino-acid (aa) residues and exhibits low degrees of homology in aa sequences in the structurally conserved region. The Spen gene was first identified in a Drosophila recessive lethal mutant 8. Orthologs were subsequently found in mice, humans and Caenorhabditis elegans 9. Studies on Drosophila melanogaster suggested that Spen mutations influenced the interactions of some signaling pathways and transcription factors related to cell development and differentiation 10, 11. Ariyoshi and Schwabe 9 speculated that the function of the SPOC domain was to facilitate protein-protein interactions in the transcription repression complex. Hiriart et al. 12 suggested that the Spen protein might mediate the export and splicing of mRNA while Sánchez-Pulido et al. 13 utilized bioinformatic methods to identify genes encoding proteins similar to Spen in protozoan and plants, and suggested that the Spen protein may play a role in programmed cell death and tumor generation, and thus providing a potential novel function for the SPOC domain.
Here we developed a promoter trap system to isolate genes specifically expressed in rice seed. In the transgenic plant 107#, the β-glucuronidase (GUS) reporter gene was expressed specifically in the endosperm. We describe the candidate gene tagged in plant 107#, termed OsRRM, and demonstrate that the protein encoded by OsRRM possesses the characteristic structure of Spen proteins. Further analysis demonstrated that OsRRM was specifically expressed in the endosperm and the ectopic expression of OsRRM caused abnormal phenotypes in transgenic plants. The function of OsRRM in the endosperm is also discussed.
Materials and Methods
Construction of the promoter trap plasmid
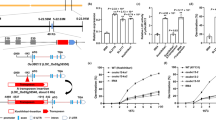
The p13GUS plasmid was obtained by inserting a 2.1-kb fragment containing the GUS gene coding region, and the nos termination region derived from vector pBI101.1 into pCAMBIA1300 between the EcoRI and HindIII sites (Figure 1A).
Schematic diagram of the T-DNA insertion site in the OsRRM gene in 107#. (A) Schematic representation of the T-DNA insertion in rice 107#. (B) Sequence of BAC OJ1381_H04 corresponding to the insertion site of T-DNA in rice 107#. (C) OsRRM gene structure from BAC OJ1381_H04. (D) Schematic representation of OsRRM coded by the cDNA (J023129A05), showing RRM, SPOC and L-zip domains. R, T-DNA right border; L, T-DNA left border; GUS, GUS reporter gene coding region; 35S, CaMV 35S promoter; HYG, hygromycin-resistant gene; ATG, translation initiation codon; TGA, termination codon; probe A and probe B, probes used for Southern hybridization in Figure 3; RRM1 and RRM2, RNA recognition motif; SPOC, Spen paralog and ortholog C-terminal domain; L-ZIP, leucine zipper-like structure. Line indicates intron; Box indicates exon.
Construction of the OsRRM-promoter∷GUS plasmid
The CaMV 35S promoter was excised from the vector pCAMBIA1301 to produce plasmid pG. A 2 kb DNA fragment upstream of the initiation codon ATG of the OsRRM gene was amplified from total DNA of Zhonghua 11 (Oryza sativa subsp. japonica cv.) by using primers 5′-GCG AGC TCT TTT TCC ATC CTT GAG-3′ and 5′-GAG GAT CCT CTG CAA GAA CCA AG-3′ with KOD PLUS DNA polymerase (Toyobo). The amplified fragment was verified by sequencing and was inserted between BamHI and SacI of pG, upstream of GUS gene, to obtain the plasmid pRMP.
Construction of the pHB-OsRRM over-expression plasmid
A 3 018 bp fragment of the OsRRM coding region was amplified from cDNA clone J023129A05 using primers 5′-ATG GGG AGA CCT CGA G-3′ and 5′-CTA CTT CTG GCC ACC AGG T-3′ with KOD PLUS DNA polymerase (Toyobo). The amplified fragment was verified by sequencing and inserted into the HindIII site, downstream from the double CaMV 35S promoter, utilizing a pHB vector (kindly provided by Professor Hongquan Yang). Insertion direction was verified by BamHI. The plasmid with consistent orientation with CaMV 35S promoter was named as pHB-OsRRM.
Construction of plasmids for GFP-fusion expression and subcellular localization examination
To construct a GFP fusion vector for onion transient transformation, a 3 015 bp fragment of the OsRRM coding region, lacking TAG, was amplified from cDNA clone J023129A05 with primers 5′-ATG GGG AGA CCT CGA G-3′ and 5′-CTT CTG GCC ACC AGG T-3′ with KOD PLUS DNA polymerase (Toyobo). The amplified fragment was verified by sequencing and inserted into SalI site between the CaMV 35S promoter and GFP coding region of the pA7-GFP vector (kindly provided by Dr K Czempinski), in-frame with GFP, resulting in pA7-OsRRM-GFP.
To construct a GFP fusion vector for rice transformation, an Ubi promoter from pHM2 (kindly provided by Professor Jingliu Zhang) was inserted into the PstI site of the pCAMBIA1300. The OsRRM∷GFP fragment from the pA7-OsRRM-GFP vector was inserted downstream of the Ubi promoter, resulting in p13U-OsRRM-GFP.
Plant transformation
Agrobacterium-mediated rice (Zhonghua 11) transformation was performed as previously described 14.
GUS assay
Histochemical GUS staining and quantitation of GUS activity was performed according to Jefferson (1987) 15.
Isolation of T-DNA flanking sequences
Sequence flanking the T-DNA right border was isolated by inverse PCR according to Ochman et al.16. Genomic DNA (1 μg) obtained from plant 107#-2 was digested with SacI, self-ligated with T4 DNA ligase (Promega), then amplified by nested PCR with the following primer sets: IR1 (5′-GAC GTA ACA TAA GGG ACT GAC CAC-3′) and SR1 (5′-GCG TTG GCG GTA ACA AGA AAG-3′); IR2 (5′-TTA CAA CGT CGT GAC TGG GAA AAC-3′) and SR2 (5′-CAA TGA ATC AAC AAC TCT CCT GG-3′); IR3 (5′-CGA ATG CTA GAG CAG CTT GAG-3′) and SR2. These sets were used in the first, second and third rounds of PCR, respectively.
Genomic sequence flanking the T-DNA left border was confirmed by PCR according to the information of the genomic sequence flanking the right border. The primers were as follows: IL1 (5′-GAG GGC AAA GAA ATA GAG TAG ATG-3′) and F1 (5′-CGT CAA CAG CAG CGG GTA GTG-3′).
Southern blot analysis
Total DNA (20 μg), isolated from leaves of plant 107# or Zhonghua 11, was digested with restriction enzymes, separated on a 0.7% agarose gel, blotted onto a nylon membrane (Amersham) and DNA detection was performed using ECL nucleic acid labeling and a detection kit (Amersham). The probe utilized was the 2.1 kb BamHI-EcoRI fragment from plasmid p13GUS (probe A in Figure 1) or the 1.9 kb PCR-amplified fragment at the 3′ end of OsRRM (probe B in Figure 1) with primers 5′-TGA GGC ATC AAA CAT GTC CTA C-3′ and 5′-AAG GGA AGT GGT ATT CAT GGG TC-3′.
Reverse transcription-polymerase chain reaction analysis
Total RNA was isolated from immature seeds 5, 10 and 20 days after pollination (DAP) of Zhonghua 11 and 107#, following Trizol (Invitrogen) instruction manual. Total RNA (1 μg) was used to generate the first strand of cDNA with the AMV Reverse Transcription Kit (Promega). Cycle conditions of PCR amplification of 1/10 volume of cDNA for desired products were as follows: initial denaturing (94 °C, 4 min), 30 cycles (denaturing, 94 °C, 45 s; annealing, 60 °C, 45 s; amplification, 72 °C, 30 s) and a final extension (72 °C, 10 min). Primer sequences were as follows: OsRRM gene: forward primer (5′-ACC GTC TGC TCA GGA AGA CCA TC-3′) and reverse primer (5′-AAG GGA AGT GGT ATT CAT GGG TC-3′); Actin gene used as control: forward primer (5′-GAA CTG GTA TGG TCA AGG CTG-3′) and reverse primer (5′-ACA CGG AGC TCG TTG TAG AAG-3′). The amplified fragment of Actin was 250 bp in size and that of OsRRM was 406 bp. All fragments were verified by sequencing.
Subcellular localization of GFP or GFP-fusion proteins in onion and rice
Onion epidermal cells were bombarded with plasmids of pA7-GFP and pA7-OsRRM-GFP using a particle gun-mediated system (Bio-Rad) 17. Bombarded tissues were incubated in the dark (12 h) before visualization of transient expression using a confocal laser-scanning microscope (Carl Zeiss LSM 510) with a standard filter set.
Zhonghua 11 calli were transformed by plasmid p13U-OsRRM-GFP as previously described 14 to obtain transgenic plants. Transgenic plant roots were also observed by laser-scanning microscopy.
Western blot assay
The EcoRI-SalI fragment (1 314 bp from aa 567 to aa 1 004aa) from OsRRM cDNA was inserted in-frame into vector pET28-A(+) (Novagen) and the His-tagged fusion peptide was expressed in E. coli BL21 (DE3), purified using Ni-NTA resin (Qiagen) and SDS-PAGE. Anti-OsRRM antibody was obtained from immunized Rabbits.
Protein gel-blot analysis was performed according to Zhang 18. Approximately 50 μg of total protein from different rice tissues, determined by Bradford dye binding assay 19 with BSA as standard, was fractionated in a 10% SDS-PAGE minigel and electroblotted to a polyvinylidene difluoride membrane (Amersham). The blot was blocked with 5% dry milk in PBST (80 mM Na2HPO4, 20 mM NaH2PO4, 100 mM NaCl and 0.1% Tween-20) and incubated with anti-OsRRM primary antibody (1:8 000), washed in PBST three times, incubated with secondary antibody (goat anti-rabbit IgG; Amersham) (1:5 000) conjugated with horseradish peroxidase then washed again. The ECLPlus System (Amersham) was used for immunodetection.
Results
Promoter trap for GUS-positive plant identification
The p13GUS plasmid, containing the promoterless GUS reporter gene in the T-DNA region (Figure 1A), was constructed to create a promoter trap system. Transformation of this plasmid into Zhonghua 11, mediated by Agrobacterium tumefaciens, resulted in 415 independent transgenic lines, of which 15 were GUS-positive. Among the GUS-positive plants, plant 107#showed blue staining only in the endosperm of T0 and succeeding generations (Figure 2A and 2B). Southern blot data suggested that a single copy of T-DNA was inserted in 107# genome (Figure 3A). Homozygous lines 107#-2 and 107#-14 were obtained by screening T2 and T3 plants with hygromycin, and homozygosity was confirmed with GUS staining (data not shown).
Southern hybridization analysis. (A) Total DNA from 107# was digested with restriction enzymes BamHI (B), EcoRI (E) and HindIII (H). GUS gene DNA fragment was used as a probe (probe A in Figure 1). (B) Total DNA from Zhonghua 11 was digested with restriction enzymes BamHI (B) and EcoRI (E). The 1.9 kb DNA sequence at the 3′ end of OsRRM was used as a probe (probe B in Figure 1).
Tagging of the candidate OsRRM gene in plant 107#
Total DNA was extracted from the homozygous line 107#-2 and the flanking sequence around T-DNA was isolated with inverse PCR. Sequencing and BLAST analyses suggested that the right flanking sequence of 800 bp was 99% identical to that in the BAC clone OJ1381_H04 while the left flanking sequence of 140 bp was 100% identical. These data suggested that the T-DNA was inserted in the chromosome region corresponding to the BAC clone OJ1381_H04 of rice chromosome 9, between positions 98448 and 98478. The 5′ end of the GUS gene in the T-DNA was close to position 98478. Because of the T-DNA insertion, a deletion of a 29 bp occurred at the insertion site of the rice chromosome (Figure 1B).
BLAST analysis also revealed that the T-DNA left flanking sequence was homologous to the sequence at the 5′ end of the cDNA clone J023129A05 (http://cdna01.dna.affrc.go.jp/cDNA). The J023129A05 cDNA sequence shows 100% identity with the corresponding sequence in the BAC clone OJ1381_H04, and encodes a protein containing two RRM domains. This trapped candidate gene was named as OsRRM, and the T-DNA was inserted within the 5′ untranslated region of OsRRM (Figure 1D).
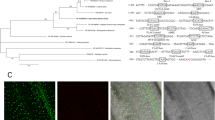
We identified five exons and four introns in OsRRM (Figure 1C), and its cDNA (3 455 bp) contains a 3 018 bp reading frame, encoding a 1 005 aa residue protein. Analysis of the protein structure showed that there are two RRM domains at the N-terminus, which could specifically bind with RNA. RRM1 is located at aa 51-116 and RRM2 is located at aa 183–238. The two RRM domains displayed 28% identity and 39% similarity, and were very similar to their counterparts in other RRM proteins such as Tial, SHARP and Spen. A leucine zipper-like structure was identified at the C-terminus, suggesting a role in protein-protein interaction and formation of protein dimers. Additionally, a conserved structural SPOC domain was identified in the middle of OsRRM (aa 498-594). In animals, RRM proteins containing SPOC domains are classified as Spen proteins. Alignment of aa sequences of the RRM and SPOC domains in OsRRM with those in other Spen proteins of other species demonstrated that (1) OsRRM shares good identity with the core region of the RRM domain in human Tia1 (37% identity and 70% similarity) and mouse Tia1 (31% identity and 45% similarity); (2) The first RRM domain of OsRRM shares good similarity with the first RRM of the Spen family members, such as HuRBP15 (53% similarity) (Figure 4A), and the second RRM domain is relatively similar to the third RRM domain of Spen members (Figure 4B); (3) The aa sequence in the mid-region of OsRRM displays good similarity with the SPOC domain at the C-terminus of Spen members (Figure 4C). Taken together, these data suggest that OsRRM is a member of the Spen gene family in rice. It is similar to FPA in Arabidopsis thaliana, in addition to other Spens in animal species (Figure 4).
Amino-acid sequence alignment of OsRRM RRM and SPOC domains. Two RRMs and one SPOC of OsRRM are aligned to the most similar domains identified in the database. Boxed regions show RNP (ribonucleoprotein domain)-2 and RNP-1 sub-domains within the RRMs. Consensus for RNP domains is defined by Burd and Dreyfuss 21. Black regions denote amino-acid residues identical to those in OsRRM, and gray regions denote similar amino-acid residues. Numbers indicate amino-acid positions. The conserved secondary structures (α, β elements) of the domains are shown below the sequence. (A) OsRRM RRM1 is aligned to the first RRM of AtFPA (A. thaliana, AY849993.1), At4g12640 (A. thaliana, AL161534.2) and HuRBP15 (Homo sapiens, NM_013286.3). (B) OsRRM RRM2 is aligned to the third RRM of AtFPA, At4g12640 and HuTIA1 (H. sapiens, NM_204356.1). (C) OsRRM SPOC is aligned to the SPOC of AtFPA, At4g12640, HuSHARP (H. sapiens, AL450998.19) and HuRBP15.
OsRRM is a single-copy gene in rice
In order to verify the copy number of OsRRM in rice, total DNA was extracted from the leaves of Zhonghua 11, digested with EcoRI and BamHI, and subjected to Southern hybridization, with a 1.9 kb sequence at the 3′ end of OsRRM used as the probe (probe B in Figure 1). Only one band was detected, suggesting that OsRRM is a single-copy gene in the rice genome (Figure 3B).
OsRRM promoter drives GUS expression in rice endosperm
To confirm that the GUS expression pattern in 107# plant was directed by the OsRRM promoter, we generated a pRMP construct by putting a 2 kb promoter region of OsRRM in front of the GUS reporter gene (Figure 2D). The calli from Zhonghua 11 were transformed, and histochemical staining of the root, stem, leaf, flower and seed of the T0 generation of transgenic plants was performed. In the 26 transgenic plants investigated, GUS staining was found selectively in the endosperm of four plants (Figure 2C) and in both the endosperm and pollen of nine plants (data not shown); there was no detectable GUS staining in the remaining 13 transgenic plants. These results, together with the GUS expression pattern in 107# plant, confirmed that the promoter of OsRRM is capable of directing specific GUS expression in the rice endosperm.
OsRRM gene expression patterns
Histochemical GUS assays carried out in 107# revealed that the GUS reporter gene was expressed specifically in the endosperm. To determine the tissue distribution of the protein encoded by the OsRRM gene, proteins were extracted from different tissues of Zhonghua 11. Equal amounts of total protein from root, stem, leaf, flower, embryo and endosperm were used for western blot analysis. A band at about 110 kDa was identified only in the endosperm sample (Figure 5), suggesting that OsRRM is specifically expressed in rice endosperm.
No detectable GUS staining was observed in 107# plant seeds before 7 DAP, whereas GUS was expressed in the whole endosperm from 7 DAP, increasing gradually and accumulating at the outer border of endosperm during later stages (Figure 2A and 2B). GUS staining was not found in the embryo during any stages of development. In addition, quantitative measurement of GUS activity was carried out in flowers and seeds of 5, 10, 15, 20, 25 and 30 DAP, with the highest GUS activity detected in seeds of 20 DAP (Figure 6).
OsRRM protein is localized to the cell nucleus
We further characterized the subcellular localization of the RNA binding protein encoded by OsRRM. The coding region of the OsRRM gene was inserted into a pA7-GFP vector and fused with the coding region of GFP to construct a plasmid (pA7-OsRRM-GFP) that was then introduced, by a particle gun, into onion epithelial cells. Visualization of transient expression using a confocal laser-scanning microscope revealed that the GFP protein was distributed throughout the cytoplasm when pA7-GFP was introduced as a control (Figure 7A–C). However, the introduction of the pA7-OsRRM-GFP plasmid resulted in the localization of OsRRM∷GFP fusion protein solely to the cell nucleus (Figure 7D–G).
Subcellular localization of the OsRRM∷GFP fusion protein. pA7-GFP transformed onion epithelial cells were observed under fluorescence light (A), or visible light (B). (C) The overlay of (A and B). Bar = 20 μm. pA7-OsRRM-GFP transformed onion epithelial cells were observed under fluorescence light (D), visible light (E) or fluorescence light after staining with DAPI (F). (G) The overlay of (D, E and F). Bar = 20 μm. Root cells of p13U-OsRRM-GFP transgenic plants were observed under fluorescence light (H), visible light (I), or fluorescence light after staining with DAPI (J). (K) The overlay of (H, I and J). Bar = 5 μm.
To determine whether OsRRM is localized to the cell nucleus in rice, the OsRRM∷GFP fusion gene was inserted downstream of the Ubi promoter to construct the plasmid p13U-OsRRM-GFP, which was then transformed into Zhonghua 11 mediated by A. tumefaciens. The OsRRM∷GFP fusion protein was found to localize to the cell nucleus in roots of transgenic plants as observed by confocal laser-scanning microscopy (Figure 7H–K), similar to the results obtained in onion cells.
Effects of OsRRM ectopic expression on rice growth
To further investigate the function of OsRRM, an OsRRM over-expression vector pHB-OsRRM (Figure 8A) controlled by double CaMV 35S promoters was constructed to transform Zhonghua 11. A total of 14 independent transgenic plants were obtained. Transgenic lines showed morphological alterations in T1 generation, with small size, late flowering, small spikes and low fructification rates (Figure 8B and 8C). However, no differences were observed in seeds between the transgenic lines and untransformed Zhonghua 11. To confirm that growth defects were caused by the over-expression of OsRRM, total proteins were extracted from leaves of transgenic lines with (lines 7 and 9) and without altered phenotypes (line 6). Western blot showed that OsRRM protein was only detectable in lines 7 and 9, but not in line 6 (Figure 8D), suggesting that ectopic expression of OsRRM resulted in abnormalities of transgenic plants.
Analyses of T1 plants transformed with the plasmid pHB-OsRRM. (A) Schematic diagram of pHB-OsRRM. (B) Phenotype of line 7 transgenic T1 plants. (C) Phenotype of spikes of line 7 transgenic T1 plants. (D) Western blot assay of seed (S) and leaf (L) of Zhonghua 11, and leaf of transgenic plant lines 6, 7 and 9.
Expression of the OsRRM gene in 107#
There were no observable phenotype alterations in 107#. To investigate whether OsRRM expression was influenced by the T-DNA insertion in 107#, RNA and protein were extracted from seeds of 107# and Zhonghua 11 for reverse transcription-polymerase chain reaction (RT-PCR) and western blot analyses. RT-PCR and sequencing data showed that the specific OsRRM band was detected in both Zhonghua 11 and 107#, while the level of OsRRM transcript was weaker in 107# than in Zhonghua 11 (Figure 9A). Similar results were obtained by western blot analysis showing that the seed OsRRM protein content of 107# was less than that of Zhonghua 11 (Figure 9B). These results suggested that although the T-DNA was inserted in the 5' UTR of OsRRM, expression of OsRRM was not completely silenced; furthermore, decreased OsRRM expression did not result in observable phenotype alterations in 107#.
Discussion
The promoter trap is a method used to identify genes based on the expression of the reporter gene, without the need for recognizable mutant phenotypes in transgenic plants. In our study, the GUS reporter gene was expressed specifically in the endosperm in a promoter trap transgenic rice plant, 107#. The T-DNA was inserted into the 5′ UTR of OsRRM in 107# and the expression pattern of OsRRM was confirmed by promoter analysis and western blot.
There were no observable phenotype alterations in 107#. The transcript and protein levels of OsRRM were analyzed in 107# and its parent Zhonghua 11 by RT-PCR and western blot. It is interesting that expression of OsRRM was not completely knocked out by the T-DNA insertion in its 5′ UTR. Similar phenomenon has been observed in other species. For example, in the study of A. thaliana frataxin mutants, Busi et al.20 found that in atfh-1 mutants, a T-DNA was located in the 5′-UTR of AtFH, and, while the T-DNA insertion strongly reduced AtFH expression, it did not completely block its transcription.
The OsRRM gene encodes a 1 005 aa protein with two RRMs at the N-terminus. RRM domains have hydrophobic aa residues of uncertain length and have a nonconserved region. It has been shown that the sequence and length of RRM domains define the RNA binding specificity of these proteins 21. Proteins containing an RRM domain have been implicated in transcription and post-transcriptional processing in the cell nucleus, as well as in the maintenance of mRNA stability, mRNA degradation and regulation of protein translation 22.
Here we demonstrate that the protein encoded by OsRRM is localized to the nucleus, suggesting a role in RNA transcription and processing. OsRRM shares good similarity with the core region of the RRM domain of Tia1 in humans or mice. Tia1 is an effector of programmed cell death and regulates development through controlling alternative splicing of the MYPT1 transcript 23. During maturation of the RNA molecule, alternative splicing of introns plays an important role in the regulation of gene expression.
In animals, RRM proteins containing SPOC domains are classified as Spen proteins. To our knowledge, this is the first report identifying OsRRM as a Spen gene in rice. BLAST analysis revealed that OsRRM is similar to FPA in A. thaliana, a component of the autonomous pathway regulating flowering time. FPA contains both RRM and SPOC domains, and is considered as the exclusive Spen gene previously reported in plants. In Arabidopsis flower development, FPA and FCA not only encode RNA binding proteins but also autoregulate gene expression through a mechanism of alternative splicing, and control FLC mRNA accumulation and flowering time 24, 25.
Endosperm cells form a tissue that is highly specialized for the rapid and efficient accumulation of storage reserves. They accumulate carbohydrates in the process of seed maturation and provide nutrition for the embryo during germination. Here, we demonstrate that OsRRM is expressed specifically in the endosperm. Studies on animals have demonstrated that Spen proteins regulate the expression of factors important in cell signaling, cell differentiation and cell fate 9, 10, 11, 12, 13. It was later found that aspects of glial development, including migration, wrapping, and survival or maintenance of the midline glial cell fate, were defective in the spen mutant of Drosophila 11.
What is the biological significance of an endosperm-specific gene in rice that harbors conservative domains similar to Spen? When the OsRRM gene was driven by the constitutive CAMV 35S promoter in transgenic rice, it was uniformly over-expressed in the whole plant, resulting in many abnormalities, such as short stature, late flowering, small spikes and low fructification rates. The morphological alterations suggested an inhibited plant growth, which may be caused by the ectopic expression of OsRRM in tissues other than endosperm. Considering the presence of specialized cells in the endosperm and the reported function of Spen genes, our data implicated OsRRM in the regulation of cell development in the endosperm.
To further investigate the function of OsRRM in rice endosperm development, we also constructed a dsRNAi plasmid to transform the calli of Zhonghua 11. This did not result in any phenotypic variations, including the size and weight of the seeds, in transgenic plants in which OsRRM expression was completely blocked. We then analyzed cooking and taste qualities, such as amylose content, gel consistency and RVA profile, in the seeds of these transgenic lines and found no difference from the untransformed Zhonghua 11 (data not shown). These data suggested that OsRRM may have a redundant function in rice that can be compensated by other homologous gene(s).
In conclusion, we have identified the first rice Spen gene, OsRRM, which is expressed specifically in the endosperm. Results of this work will facilitate a greater understanding of the function of OsRRM in rice endosperm development. However, further research is needed to fully understand the cell types or signal pathways affected by over-expression of OsRRM in plants during the seedling stage, and to elucidate the exact biological function and molecular mechanisms of OsRRM in the endosperm.
Abbreviations
- (GUS) :
-
β-glucuronidase
- (RRM):
-
RNA recognition motif
- (SPOC):
-
Spen paralog and ortholog C-terminal
- (Spen) :
-
split ends
- (RT-PCR):
-
reverse transcription-polymerase chain reaction
- (DAP):
-
day after pollination
- (RNP):
-
ribonucleoprotein domain
References
Berger F . Endosperm: the crossroad of seed development. Curr Opin Plant Biol 2003; 6:42–50.
McLaughlin M, Walbot V . Cloning of a mutable bz2 allele of maize by transposon tagging and differential hybridization. Genetics 1987; 117:771–776.
Jonsson JJ, Weissman SM . From mutation mapping to phenotype cloning. Proc Natl Acad Sci USA 1995; 92:83–85.
Li X, Zhang YL . Reverse genetics by fast neutron mutagenesis in higher plants. Funct Integr Genomics 2002; 2:254–258.
Osborne BI, Baker B . Movers and shakers, Maize transposons as tools for analyzing other plant genomes. Curr Opin Biol 1995; 7:406–413.
An G, Lee S, Kim SH, Kim SR . Molecular genetics using T-DNA in rice. Plant Cell Physiol 2005; 46:14–22.
Springer PS . Gene traps: tools for plant development and genomics. Plant Cell 2000; 12:1007–1020.
Kolodziej PA, Jan LY, Jan YN . Mutations that affect the length, fasciculation, or ventral orientation of specific sensory axons in the Drosophila embryo. Neuron 1995; 15:273–286.
Ariyoshi M, Schwabe JW . A conserved structural motif reveals the essential transcriptional repression function of Spen proteins and their role in developmental signaling. Genes Dev 2003; 17:1909–1920.
Wiellette EL, Harding KW, Mace KA, Ronshaugen MR, Wang FY, McGinnis W . Spen encodes an RNP motif protein that interacts with Hox pathways to repress the development of head-like sclerites in the Drosophila trunk. Development 1999; 126:5373–5385.
Chen F, Rebay I . Split ends, a new component of the Drosophila EGF receptor pathway, regulates development of midline glial cells. Curr. Biol 2000; 10:943–946.
Hiriart E, Gruffat H, Buisson M, et al. Interaction of the Epstein-Barr virus mRNA export factor EB2 with human Spen proteins SHARP, OTT1, and a novel member of the family, OTT3, links Spen proteins with splicing regulation and mRNA export. J Biol Chem 2005; 280:36935–36945.
Sánchez-Pulido L, Rojas AM, van Wely KH, Martinez-A C, Valencia A . SPOC: A widely distributed domain associated with cancer, apoptosis and transcription. BMC Bioinformatics 2004; 5:91.
Liu QQ, Zhang JL, Wang ZY, Hong MM, Gu MH . A highly efficient transformation system mediated by Agrobacterium tumefaciens in rice. Acta Phytophsiol Sin 1998; 24:259–275 (in Chinese).
Jefferson RA . Assaying chimeric genes in plants: the GUS fusion system. Plant Mol Biol Rep 1987; 5:387–405.
Ochman H, Gerber AS, Hartl DL . Genetic applications of an inverse polymerase chain reaction. Genetics 1988; 120:621–623.
Kuijt SJ, Lamers GE, Rueb S, et al. Different subcellular localization and trafficking properties of KNOX class 1 homeodomain proteins from rice. Plant Mol Boil 2004; 55:781–796.
Zhang YC, Gong SF, Li QH, Sang Y, Yang HQ . Functional and signaling mechanism analysis of rice Cryptochrome 1. Plant J 2006; 46:971–983.
Bradford MM . A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976; 72:248–254.
Busi MV, Maliandi MV, Valdez H . Deficiency of Arabidopsis thaliana frataxin alters activity of mitochondrial Fe-S proteins and induces oxidative stress. Plant J 2006; 48:873–882.
Burd CG, Dreyfuss G . Conserved structures and diversity of functions of RNA-binding proteins. Science 1994; 265:615–621.
Birney E, Kumar S, Krainer AR . Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res 1993; 21:5803–5816.
Shukla S, Dirksen WP, Joyce KM, Guiner-Blanvillain CL, Breathnach R, Fisher SA . TIA proteins are necessary but not sufficient for the tissue-specific splicing of the myosin phosphatase targeting subunit 1. J Biol Chem 2004; 279:13668–71366.
Schomburg FM, Patton DA, Meinke DW, et al. FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. The Plant Cell 2001; 13:1427–1436.
Quesada V, Dean C, Simpson GG . Regulated RNA processing in the control of Arabidopsis flowering. Int J Dev Biol 2005; 49:773–780.
Acknowledgements
We are grateful to Qiao-quan Liu for technical assistance. We also thank Lin-sheng An for planting the transgenic plants. This work was supported by grants from the Ministry of Science and Technology of China (No. 2006AA10A102 and No. 2005CB120803) and the National Natural Science Foundation of China (No. 30671112).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, SY., Wang, ZY. & Cai, XL. OsRRM, a Spen-like rice gene expressed specifically in the endosperm. Cell Res 17, 713–721 (2007). https://doi.org/10.1038/cr.2007.43
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2007.43
Keywords
This article is cited by
-
Genetic differentiation and restricted gene flow in rice landraces from Yunnan, China: effects of isolation-by-distance and isolation-by-environment
Rice (2021)
-
Transcript analysis of abscisic acid-inducible genes in response to different abiotic disturbances in two indica rice varieties
Theoretical and Experimental Plant Physiology (2019)
-
Comparative Analysis of the Expression of Candidate Genes Governing Salt Tolerance and Yield Attributes in Two Contrasting Rice Genotypes, Encountering Salt Stress During Grain Development
Journal of Plant Growth Regulation (2019)
-
Transcriptome Profiling of Abiotic Stress-Responsive Genes During Cadmium Chloride-Mediated Stress in Two Indica Rice Varieties
Journal of Plant Growth Regulation (2018)
-
Functional conservation of MtFPA, a nucleus-localized RNA-recognition motif-binding protein that regulates flowering time in Medicago truncatula
Plant Biotechnology Reports (2018)