Abstract
Parthenogenesis is one of the main, and most useful, methods to derive embryonic stem cells (ESCs), which may be an important source of histocompatible cells and tissues for cell therapy. Here we describe the derivation and characterization of two ESC lines (hPES-1 and hPES-2) from in vitro developed blastocysts following parthenogenetic activation of human oocytes. Typical ESC morphology was seen, and the expression of ESC markers was as expected for alkaline phosphatase, octamer-binding transcription factor 4, stage-specific embryonic antigen 3, stage-specific embryonic antigen 4, TRA-1-60, and TRA-1-81, and there was absence of expression of negative markers such as stage-specific embryonic antigen 1. Expression of genes specific for different embryonic germ layers was detected from the embryoid bodies (EBs) of both hESC lines, suggesting their differentiation potential in vitro. However, in vivo, only hPES-1 formed teratoma consisting of all three embryonic germ layers (hPES-2 did not). Interestingly, after continuous proliferation for more than 100 passages, hPES-1 cells still maintained a normal 46 XX karyotype; hPES-2 displayed abnormalities such as chromosome translocation after long term passages. Short Tandem Repeat (STR) results demonstrated that the hPES lines were genetic matches with the egg donors, and gene imprinting data confirmed the parthenogenetic origin of these ES cells. Genome-wide SNP analysis showed a pattern typical of parthenogenesis. All of these results demonstrated the feasibility to isolate and establish human parthenogenetic ESC lines, which provides an important tool for studying epigenetic effects in ESCs as well as for future therapeutic interventions in a clinical setting.
Similar content being viewed by others
Introduction
Human embryonic stem cells (hESCs) have tremendous potential for cell therapy of human diseases such as neurondegenerative disorders 1, and in regenerative medicine 2. Since the first derivation of human embryonic stem cell lines from IVF blastocysts 3, the field of hESC research has generated substantial interest, although certain obstacles still remain including limited sources of oocytes and controversial ethical issues 4 that have hindered further advancement. Parthenogenetic stem cells are regarded as a substitute for ES lines derived from somatic cell nuclear transfer (SCNT), with higher efficiency and less ethical controversy, and proven results in mouse and non-human primate models 5, 6.
Parthenogenetic stem cell lines have been extensively reported in mouse 7 and non-human primate models 8, 9 and recently in human 10, 11. Parthenogenetic stem cells have been successfully differentiated into neuronal cells in a defined differentiation system in monkey in vitro8, and another report showed that dopamine neurons derived from parthenogenetic ESCs have long term survival after transplantation into rodent and primate brains 6. For patients with Parkinson's or other neurodegenerative disorders (especially in female patients), there may be hope for autologous and personalized stem cell treatments of the otherwise devastating diseases. However, one report has suggested that the loss of epigenetic imprinting may limit the effectiveness of stem cells derived from parthenogenetic embryos 12.
As paternally imprinted genes were shown to be stable in androgenetic (two paternal chromosome sets) stem cells 13, maternal factors seem to have a major effect on the instability of imprinted genes 14. In parthenotes all genetic material originates from the maternal genome, and the resulting stem cells would demonstrate only maternal patterns of gene imprinting. Thus, parthenogenetic ESCs can be an invaluable source of material to study mechanisms of epigenetic regulation, as well as to explore disease mechanisms and their treatment, since many severe diseases including cancer 15 and Beckwith-Wiedeman syndrome 16 are believed to be influenced by imprinting.
In this study, we reported the establishment of two human embryonic stem cell lines from parthenogenetically activated embryos in vitro and subsequent observation of those cells over more than 100 passages. We verified the parthenogetic origin of these cell lines and performed detailed characterization of the pluripotency of one of the lines.
Materials and Methods
Informed consent for egg donation
All of the egg donors were on the assisted reproductive technology (ART) cycle. No financial benefit was involved in the donation process. The egg donors were informed of all details of the procedure, including egg utility and research destination, and signed a series of informed consent documents voluntarily. We guaranteed that all of the eggs were used in basic scientific research, and not for reproductive purposes.
All of the ART and therapeutic cloning procedures followed closely the guidelines legislated and posted by the Ministry of Health of the People's Republic of China. Normally only 1-4 eggs were received and selected randomly from each donor, so that it would not jeopardize the future clinical treatment or reproductive capability of the donors.
Donor superovulation and oocyte collection
Before subject to superovulation, each patient underwent basic physical examination and was tested to rule out human immunodeficiency virus (HIV), hepatitis B virus (HBV), hepatitis C virus (HCV) and contagious venereal diseases. Eligible patients received 1.3 mg GnRH-agonist (Dapherin) at mid-luteal phase and recombined follicle stimulating hormone (rFSH) (gonal-f, Serono, Sweden) at a dose of 150-300IU/day from day 3 of menstrual cycle for the growth of multiple follicles. When the diameter of dominant follicles reached 18 mm, a single 10 000 IU dose of HCG (Profasi, Serono, Sweden) was administered and transvaginal follicular aspiration was performed 36 h later.
Parthenogenetic activation and embryo culture
Donated eggs were selected randomly, and cultured in a humidified atmosphere at 37 °C with gas mixture consisting of 90% N2, 5% O2 and 6% CO2. Prior to activation, granule cells around individual oocytes were removed by brief exposure to 80 IU/ml hyaluronidase (Sigma) (∼ 30 s) followed by sequential mechanical manipulation in HEPES buffered medium (Quinn'S AdvanlageTM Manipulation Medium). Mature oocytes were subjected to the following activation procedure, and immature oocytes were cultured in cleavage medium (Quinn'S AdvanlageTM Cleavage Medium). After culturing for 30 min in a humidified atmosphere at 37 °C with gas mixture (90% N2, 5% O2 and 6% CO2), mature oocytes were subjected to sequential activation treatments. First, electric activation was performed with electric pulse in electrofusion medium containing 0.3 M mannitol (with 0.1 mM MgSO4·7H2O, 0.1 mg/ml polyvinyl alcohol, 3 mg/ml human serum albumin, 0.05 mM CaCl2·2H2O). After rinsing in HEPES buffer medium (Quinn'S AdvanlageTM Manipulation Medium), the oocytes were transferred into 5 μM ionomycin (Sigma) for 5 min and then incubated in cleavage medium (Quinn'S AdvanlageTM Cleavage Medium) with 2 mM 6-dimethylamino-purine for an additional 5 h. The activated oocytes were cultured in the cleavage medium (Quinn'S AdvanlageTM Cleavage Medium); pronuclear cells were observed with an inverted microscope (Nikon Co.) 18 h later and the embryos were transferred into blastocyst culture medium (Quinn'S AdvanlageTM Blastocyst Medium) two days later for another 3-4 days. The embryos were observed every 24 h and developmental efficiency was calculated.
Human stem cells isolation and culture
Embryos developed to blastocyst stage at the fifth day after activation; one or two days later, the blastocysts reached the hatching stage and an immunosurgery method was used to isolate the inner cell mass (ICM) as previously described 17. Specifically, the zona pellucida was removed by 0.5% pronase A (Sigma) treatment, and the blastocyst was incubated with horse antiserum to human spleen cells followed by exposure to guinea pig complement. Trophectoderm cells were detached from the ICM by gently pipetting the zona-free blastocysts. The isolated ICMs were plated on the mitomycin-C irradiated mouse embryonic fibroblast cells (MEFs) for about 5-9 days and primary ESC colonies were then dissociated mechanically and cultured in human ES cell culture medium consisting of 80% Knockout-DMEM (Gibco), 20% Serum-Replacement (Gibco), 1 mM glutamine (Sigma), 1% nonessential amino acid (Gibco), 0.1 mM 2-mercaptoethanal (Sigma), 50 UI/ml penicillin (Sigma), and 50 UI/ml streptomycin (Sigma) mixture as previously described 18. Human ES cell lines established from IVF embryos were used as controls. One of these IVF lines (IVF-1) was fully characterized and reported previously 18.
The primary colonies were cultured for 5-9 days and the ES cells were selected, picked up, and replated on new feeder cells. When the ES cells appeared to proliferate stably, the ES colonies were dissociated every 4-5 days by mechanical or digestive methods. All of the ESCs were cultured under 37 °C, 5% CO2 in a humidified atmosphere.
Characterization of human parthenogenetic ES cell lines
For immunoflourescence staining and assaying of alkaline phosphatase (AP), hPES cells were plated on glass coverslips coated by MEF cells and cultured for 3-4 days. The ES cell clones were fixed by acetone, and blocked with 10% goat serum in PBS (Sigma) for 1 h, and then incubated with primary antibodies overnight at 4°C. Antibody localization was performed using rabbit anti-mouse immunoglobulin secondary antibody conjugated to flourescein isothiocyanate. Specimens were observed with a fluorescent microscope. The primary antibodies included antibodies to stage-specific embryonic antigen 3 (SSEA-3), SSEA-4, SSEA-1 (Developmental Studies Hybridoma Bank at the University of Iowa), TRA-1-60 (Chemicon), and TRA-1-81 (Chemicon). Nuclei of ESCs were stained with Hoechest 33342 (Sigma).
Alkaline phosphatase (AP) activity was observed by the Gomori modified method. The ES clones were fixed by cold acetone for 10 min and incubated with AP incubation medium containing 2% sodium glycerophosphate, 2% barbital sodium, 2% chloratum kalium and 2% magnesium sulfate for 3 h at 37 °C, and subsequently incubated in 5% cobaltous nitrate for 5 min and in 1% ammonium sulfide for 1 min.
Gene expression profile analysis
Expression of imprinted genes of hPES cells was analyzed by RT-PCR. Total RNA was extracted using TRIzol (Invitrogen, Grand Island, NY). cDNA was synthesized from ∼ 1 mg of total RNA using SuperScript II reverse transcriptase (Invitrogen, Grand Island, NY) and was subjected to PCR amplification with primers for Oct-4, Rex1, Sox2, Nanog, NF68KD, α-fetoprotein, albumin, H19, h-Ube3A, h-Snprn, and Igf-2 (Table 1). β-actin was used as a control. PCR products were size-fractionated by 1% agarose gel electrophoresis and visualized by ethidium bromide staining. Final analysis was obtained in an image analyzer (Biorad).
Karyotyping
hPES colonies were incubated with 0.2 μg/ml colchicin (Invitrogen) for 2 h at 37 °C. Cells were collected and trypsinized, washed with PBS, and incubated with 0.075 mol/L potassium chloride for 10 min at 37 °C. Cells were fixed with methanol:glacial acetic acid (1:3) three times and dropped onto glass slides. Chromosome spreads were Giemsa banded and photographed. The karyotypes of ES cells were detected every 10 passages. Karyotypes were assessed by normal G-banding procedure and 50 metaphase II spreads were examined for each sample, and a normal karyotype would show normal chromosome numbers and G-banding patterns in the spreads examined.
Assessment of the differentiation capacity in vitro and in vivo
Embryoid body (EB) method was used to assess the differentiation ability in vitro of hPES cells via specific gene expression. hPES cells were removed from culture dishes using 1 mg/ml collagenase IV and cultured in suspension condition. Spontaneous EBs were grown in medium consisting of 80% Knockout-DMEM, supplemented with 20% serum replacement, 0.1 mM 2-mercaptoethanal, and 1% nonessential amino acids. After 5-14 days, all of the differentiated samples from EBs were collected, and RT-PCR was performed to analyze specific gene expression for the three embryonic germ layers–mesoderm, endoderm, and ectoderm.
Teratoma production was used to assess the differentiation ability in vivo of hPES cells. About 5-6 million undifferented hPES cells from passages 10, 15, and 20 were harvested and injected into the back muscle of 6-week-old male SCID-mice. After 3 months, the mice were sacrificed. Various tissues from teratoma were dissected, fixed in Bouins overnight, processed, and sectioned according to standard procedures 19 and counterstained with either hematoxylin and eosin or Weigert's stain. Sections were examined using bright field light microscopy and photographed as appropriate.
STR Typing
Genomic DNA samples were extracted from the ES cells, peripheral blood of oocyte donors, the teratomas using phenol-chloroform method 20. Sixteen STR loci were co-amplified in one PCR reaction. Alleles of STR loci were distinguished by fluorescence detection following electrophoretic separation. STR typing process was performed according to manufacturer's recommendations using the PowerePlEX 16 system (Promega, USA). The ABI PRISM 310 Genetic Analyzer was used for fluorescence detection of amplified fragments.
SNP analysis
SNP genotyping
Genomic DNA samples were purified from cell lines using phenol/chloroform extraction and ethanol precipitation, and submitted to the Penn Microarray Facility (University of Pennsylvania, Philadelphia PA) for genotyping. Target DNA preparation and microarray processing for the Affymetrix 250K Sty SNP Mapping GeneChips (Affymetrix, Santa Clara, CA) were conducted according to the manufacturer's protocol (Mapping 500K Assay Manual, http://www.affymetrix.com). Briefly, after digestion of genomic DNA with StyI, complexity reduction PCRs produced over 100 μg of target DNA per sample, which was fragmented by DNase treatment and biotinylated by terminal transferase. Microarrays were hybridized overnight, then washed and stained using FS450 Fluidics Stations and scanned in a GeneChip 3000 7G scanner.
SNP data analysis
The Affymetrix GCOS package with BRLMM algorithm was used to assign genotype calls for 238 304 SNPs distributed throughout the genome with median intermarker distance of 5.2 kb (mean 12.2 kb). Tabulated calls were ordered by SNP physical map coordinate, and the number of heterozygous calls was summed within a window of 1 000 contiguous SNPs. This value was divided by the DNA length in megabase-pairs encompassed by the window, producing a normalized heterozygosity rate suitable for comparison within and between chromosomes. The average heterozygosity rate for every 5 Mb interval distal from the centromere was calculated for each chromosome and color coded to represent differences in levels among samples.
Results
Parthenogenetic embryo development and derivation of ES cell lines
Nineteen oocytes prepared shortly after donation by 10 ART patients were artificially activated in vitro. After removal of cumulus cells only matured metaphase II oocytes with distinct first polar bodies were taken for a two-step activation procedure. Electric-stimulation and chemical agent stimulation were performed sequentially. Sixteen oocytes were activated and pesudo-pronuclei were observed after an additional 18 h in culture. Fourteen activated oocytes cleaved but the developmental ability thereafter decreased significantly resulting in four blastocysts after culturing for 5-6 days (Table 2). The surviving blastocysts were able to expand and hatch similar to normal fertilized embryos (Figure 1).
Three parthenogenetic blastocysts had visible ICM, and were used for derivation of ESC lines. One of the three ICMs was lost at day 3; the other two were capable of growing and were mechanically dissected after 7-9 days, and the resulting cell lines were named hPES-1 and hPES-2. Characterization and identification of these ESC lines were performed after primary hPES cells were propagated for more than 10 passages (Table 3). We will describe the complete analysis for the hPES-1 line in this paper as an example.
Characteristics of hPES cells
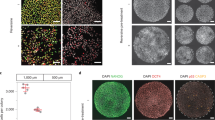
hPES colonies demonstrated stem cell morphology, and displayed traits of a normal stem cell line, including high nuclear/cytoplasm ratio, and tightly packed colonies when observed under the inverted microscope (Figure 2 and Table 4). In addition, these colonies showed high levels of AP activity (Figure 2 and Table 4). The hPES colonies expressed stem cell positive cell surface markers including SSEA-3, SSEA-4, TRA-1-60, and TRA-1-81, but not the negative cell surface markers, including SSEA-1, as expected (Figure 2 and Table 4).
Morphology, AP and Immonustaining of stem cell markers for hPES-1 cell line. (A) Morphology of hPES-1 colony under invert microscope; (B) AP staining of hPES-1; (C) SSEA4; (D) SSEA3; (E) TRA-1-60; (F) TRA-1-81; (G) SSEA1. (C–G) Listed are optical, confocal images and the corresponding Hoechst staining for the hPES-1 cells.
Expression of important pluripotency related genes was detected by RT-PCR and positive signals for Oct-4, Rex1, Sox2 and Nanog were shown in both hPES cell lines, as well as in the ESCs derived from IVF embryos (Figure 2 and Table 4).
Differentiation potential of hPES cells
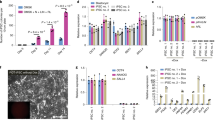
hPES cells were capable of differentiating to EB in vitro when cultured in suspension conditions. After spontaneous differentiation for 10-15 days, these EBs were used for further RT-PCR analysis. Genes uniquely expressed in all three embryonic germ layers were shown to be expressed, including albumin (endoderm), α-fetoprotein (endoderm), Glu2 (endoderm), α-cardiac actin (mesoderm) and neurofilament (ectoderm) (Figure 3).
RT-PCR analysis of EBs of hPES cells for differentially expressed genes from three embryo germ layers in vitro. Data from the hPES-1 line are shown: Lanes: 1: Albumin; 2: a-cardiac actin; 3: a-fetoprotein; 4: Glu2; 5: GAPDH; 6: NF-68KD. No primer and no template PCR controls were negative (data not shown)
Differentiation ability of hPES cells in vivo was evaluated by teratoma formation. 5-6 × 105 stem cells were injected into the back muscle of SCID-mice. While hPES-2 cells failed to form any teratoma, teratomas were observed after 3 months with cells from hPES-1 cell line (of different passages). Histopathology of the resulting teratomas revealed the presence of various tissues including glandular and intestinal epithelia (endoderm), cartilage and interstitial cells (mesoderm), and squamous cells (ectoderm) (Figure 4). These results demonstrated that hPES-1 was capable of differentiation in vivo into all three embryonic germ layers, and thus all cell types in human.
Identification of parthenogenetic origin
In normal fertilized embryos differential expression occurs in maternally or paternally imprinted genes. Therefore the lack of paternal imprint patterns would be useful evidence of parthenogenetic origin. Expression of imprinted genes was tested in both hPES cell lines and IVF control lines. Transcription of paternally expressed SNRPN and IGF-2 genes was not detected, whereas maternally expressed UBE3A and H19 were observed in hPES-1 (Figure 5). The expression of all of the above imprinted genes was detected in IVF controls.
RT-PCR analysis of the expression of selected imprinted gene expression. Lanes 1-5 are results from hPES-1 cells and lanes 6-10 normal IVF-1 hES cells for the following genes: lanes 1 and 10: β-actin; lanes 2 and 6: h-SNRPNR; lanes 3 and 7: IGF-2; lanes 4 and 8: UBE3A; lanes 5 and 9: H19. No primer and no template PCR controls were netative (data not shown).
Karyotyping was performed at every 20 passages from passages 20 to 120 for hPES-1 and hPES-2. The 46 XX karyotypes were consistent with the hPES origin from a female donor; P 40 data from hPES-1 is shown in Figure 6. No chromosomal abnormality was identified beyond more than 80 passages in hPES-1, indicating that hPES-1 sustained genetic stability through long and repeated propagation in vitro. For hPES-2, abnormal karyotypes (e.g. chromosome translocation) were observed more and more frequently beyond passage 50.
To further confirm the autologous origin, an STR study was performed among the hPES lines, lymphocytes from the donors, and the resulting teratoma from hPES-1. Results from 16 DNA loci indicated that hPES-1 was genetically identical to the teratoma and more than 99.85% comparable with the lymphocytes from the oocyte donor for hPES-1 (Table 5).
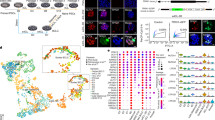
Using the Affymetrix GeneChip Human Mapping 250 K SNP Array, we extensively analyzed the patterns of heterozygosity of hPES and the IVF control lines. Heterozygosity rates for every 5 Mb interval distal from the centromeres were calculated and plotted for each chromosome of hPES and IVF control lines at early (P 12), medium (P 40) and late passages (P>70). We described the hetrozygosity rate using six arbitrary grades to identify the areas along the chromosome that are more heterozygous in nature.
The pattern of SNP genotypes of hPES-1 revealed that homozygosity predominates at pericentromeric markers and heterozygosity is seen at more distal markers (Figure 7), which resembles very closely the result described recently by Kim et al. 11. In contrast, heterozygosity was observed throughout all chromosomes with all markers with the IVF control line (Figure 7). These data strongly confirmed the parthenogenetic origin of the hPES-1 cell line. Similar results were found for hPES-2.
Genome-wide SNP Analysis on hES cells using Affymetrix 250K SNP Array. Top: Heterozygosity rate plots for representative chromosomes 1, 11, 13 and 18, with IVF-1 and hESP-1 lines at early (P 12 or P 13) and medium passages (P 39 or P 40). Heterozygosity rate is defined in Materials and Methods section. Bottom: Heterozygosity maps for the four samples above at 5 Mb resolution along each chromosome. Heterozygosity rate is indicated by color coding using six arbitrary levels: red, orange, yellow, light blue, blue and dark blue correspond to rate thresholds of 3, 6, 9, 14, 19, 25 and over.
Discussion
Two human parthenogenetic stem cell lines were successfully derived from parthenogenetic embryos. After artificial activation by a two-step procedure, 4 out of 19 initial oocytes developed into blastocysts. Two ESC lines were obtained. We employed a battery of tests to characterize the pluripotent nature of the ESCs and confirmed their parthonogenetic origin. The characteristic was reflected by the hPESs displaying the typical ESC morphology and expression of known human ESC positive markers (SSEA-4, SSEA-3, TRA-1-60, TRA-1-80), and lack of the human ESC negative markers (e.g. SSEA-1) 21, 22.
Self-renewal and pluripotency are the two important features of embryonic stem cells and are reflected in the hPES-1 line as similar proliferation time was maintained during propagation (data not shown), and genetic stability was also proven to be preserved via karyotype analysis. Pluripotency of ESCs is critical for ESC-based therapy. RT-PCR results revealed the expression of key genes such as Oct-4, Rex1, Sox2 and Nanog that coordinately play central roles suggesting, on a molecular level, the pluripotency of the hPES lines.
Another crucial function for ESCs and therefore for therapeutic cloning is the differentiation potential of the human ESCs. hPES-1 is capable of differentiation into all three germ layers in vitro and in vivo as measured by RT-PCR from differentiated EBs and histopathology analysis in resulting teratoma. hPES-2, on the other hand, expressed marker genes from all three germ layers from EBs, but failed to form teratoma in vivo. Uniparental ESCs were shown to be able to contribute to proliferating stem cell populations of postnatal aggregation chimeras 23, and differentiation potential of parthenogenetic ESCs was demonstrated in non-human primate models using EB and teratoma analysis 6, 9. A common problem for parthenogenetic ESCs is the capacity for further differentiation of these cells in vivo. Our hPES-1 and hPES-2 demonstrated significant differences in differentiation abilities in vivo. Interestingly, Hikichi et al. reported improving parthenogentic ESCs' differentiation potential using a nuclear tranasfer (NT) technique in mice 24. Thus, the potential of the parthenogentic ESCs might be enhanced by NT and other culture techniques to further our understanding and utilization of these cells in stem cell based therapy in human. In addition, while hPES-1 maintained normal karyotypes beyond 80-100 passages, hPES-2 failed to sustain genomic stability. This may suggest micro-abnormalities or other pre-existing factors/conditions in this ES cell line that hindered further ES cell functions (i.e. self renewal and pluripotency). Thus the two parthenogentic ES cell lines generated here would provide a useful model to study the mechanisms of ESC differentiation and ESC functions.
The mechanism of genomic imprinting is at present not fully understood. The uniparental ESC system provides a useful tool for the studying of epigenetic modifications and imprinting mechanisms such as allele-specific DNA methylation and chromatin structural modifications 14. In the process of parthenogenesis, the haploid genome was copied to diploid without expelling the polar body (PB), therefore all of the genetic material in the parthenote came from the maternal genome, and lacked paternal imprinting. SNRPN (small nuclear ribonucleoprotein polypeptide N) is a mono-allelic paternally expressed gene and has previously been shown to be functionally imprinted in both mouse 25 and human 26. Another paternally expressed imprinted gene, IGF2 is regarded as a key regulator for fetal growth control, and is thought to be closely linked to diseases including growth disorders and cancers 27. UBE3A and H19 genes, on the other hand, have been identified as maternally expressed imprinted genes in human, and as also regarded to contribute to human diseases and tumor growth 28, 29. A negative RT-PCR result for the two paternally expressed imprinted genes together with a positive result for UBE3A and H19 in hPESs is consistent with a parthenogenesis origin for the hPES cell lines. Failure of development to term of parthenogenetic embryos was thought to be a consequence of having a uniparental genome 30, and although parthenogenetic stem cells were suggested to have limited differentiation potential due to the lack of paternal imprinting 30, 31, modification of specifically imprinted genes was reported to help extend full organismal development and increase the differentiation ability of the ESCs and thus the ESCs-based cell therapy 32, 33, 34.
STRs (short tandem repeat) are short sequences of DNA with high frequency tandem repeats of 3-7 base pairs, and have been used extensively for parentage testing, forensic identification, and medical diagnostics 35, due to their high polymorphic rates and stable inheritance. STRs are considered an accurate, highly discriminating, sensitive, and rapid technique for DNA typing and genotype identification. Thus, we analyzed the STR polymorphisms of our hPES cells and their egg donors using the PowerPlex® 16 System (Promega). Using an accepted positive threshold for parentage identification 36, our result confirmed that the parthenogenetic ESCs truly originated from the egg donors. This result is consistent with those found by Kim et al. 5, that contrary to the presumption of predominant homozygosity in the pES cells, a higher level of heterozygosity was observed due to recombination events prior to the generation of the parthenogenetic ES lines. Nevertheless, the STR confirmed the origin of the derived parthenotes.
A recent report analyzed a human ES cell line initially thought to be generated by SCNT 37, and showed that this line was a result of parthenogenesis 11. Using high resolution single nucleotide polymorphism (SNP) analysis, Kim et al. demonstrated distinct signatures of genetic recombination that distinguish parthenogenetic ES cells from those generated by SCNT. A genome-wide SNP analysis was used as an independent verification of the parthenogenetic nature of the hPES-1 and hPES-2 stem cell lines, and our results resemble closely the parthenogenetic ES cell pattern previously reported. Moreover, while the SCNT-hES-1 line previously reported 37 was not an intentional product of parthenogenesis, our study provides a reliable method for producing and analyzing true human parthenogenetically derived ESC lines since parthenogenesis is still to date more efficient than SCNT to generate human ES cells for cell therapy.
The parthenogenetic stem cells have been shown to be capable of contributing to several mature tissues in chimeric mice 38, 39. For female patients, it is extremely attractive to use parthenogenetic embryonic stem cells for autologous cell therapy, since ethical concerns and the hazards for collecting and destroying one's normally competent embryos for ES cell derivation can be avoided. Thus, parthenogenetic ES cells have a tremendous potential for cell therapy in clinics and we offer here a useful model and an experimentally verified system for using these cells to study stem cell biology (including epigenetic regulation of ESCs) and explore their potential in disease therapeutics.
References
Lindvall O, Kokaia Z . Stem cells for the treatment of neurological disorders. Nature 2006; 441:1094–1096.
Vats A, Bielby RC, Tolley NS, Nerem R, Polak JM . Stem cells. Lancet 2005; 366:592–602.
Thomson JA, Itskovitz–Eldor J, Shapiro S, et al. Embryonic stem cell lines derived from human blastocysts. Science 1998; 282:1145–1147.
Steinbrook R . Egg Donation and Human Embryonic Stem-Cell Research. N Engl J Med 2006; 354:324–326.
Kim K, Lerou P, Yabuuchi A . Histocompatible Embryonic Stem Cells by Parthenogenesis. Science 2006; 315:482–486.
Sánchez-Pernaute R, Studer L, Ferrari D, et al. Long-term survival of dopamine neurons derived from parthenogenetic primate embryonic stem cells (cyno-1) after transplantation. Stem Cells 2005; 23:914–922.
Kaufman MH, Robertson EJ, Handyside AH, Evans MJ . Establishment of pluripotential cell lines from haploid mouse embryos. J Embryol Exp Morphol 1983; 73:249–261.
Cibelli J, Grant K, Chapman K, et al. Parthenogenetic stem cells in nonhuman primates. Science 2002; 295:819.
Vrana K, Hipp J, Goss A, et al. Nonhuman primate parthenogenetic stem cells. Proc. Natl. Acad. Sci. U S A 2003; 100:11911–11916.
Revazova ES, Turovets NA, Kochetkova OD, et al. Patient-Specific Stem Cell Lines Derived from Human Parthenogenetic Blastocysts. Cloning and Stem Cells 2007; 9:1–9.
Kim K, Ng K, Rugg-Gunn P, et al. Recombination Signatures Distinguish Embryonic Stem Cells Derived by Parthenogenesis and Somatic Cell Nuclear Transfer. Cell Stem Cell 2007; 1:346–352.
Hernandez L, Kozlov S, Piras G, Stewart CL . Paternal and maternal genomes confer opposite effects on proliferation, cell-cycle length, senescence, and tumor formation. Proc Natl Acad Sci USA 2003; 100:13344–13349.
Mann JR, Stewart CL . Development to term of mouse androgenetic aggregation chimeras. Development 1991; 113:1325–1333.
Allen ND, Barton SC, Hilton K, Norris ML, Surani MA . A functional analysis of imprinting in parthenogenetic embryonic stem cells. Development 1994; 120:1473–1482.
Feinberg AP, Ohlsson R, Henikoff S . The epigenetic progenitor origin of human cancer. Nat Rev Genet 2006; 7:21–33.
Prawitt D, Enklaar T, Gartner-Rupprecht B, et al. Microdeletion and IGF2 loss of imprinting in a cascade causing Beckwith-Wiedemann syndrome with Wilms' tumor. Nat Genet 2005; 37:785–786.
Solter D, Knowles BB . Immunosurgery of mouse blastocyst. Proc Natl Acad Sci USA 1975; 72:5099–5102.
Li T, Zhou CQ, Mai QY, Zhuang GL . Establishment of human embryonic stem cell line from gamete donors. Chin Med J 2005; 118:116–122.
Klimanskaya I, Chung Y, Becker S, Lu SJ, Lanza R . Human embryonic stem cell lines derived from single blastomeres. Nature 2006; 444:481–485.
Parayre S, Falentin H, Madec MN, et al. Easy DNA extraction method and optimisation of PCR-Temporal Temperature Gel Electrophoresis to identify the predominant high and low GC-content bacteria from dairy products. J Microbiol Methods 2007; 69:431–441.
Loring JF, Rao MS . Establishing standards for the characterization of human embryonic stem cell lines. Stem Cells 2006; 24:145–150.
Brivanlou AH, Gage FH, Jaenisch R, Jessell T, Melton D, Rossant J . Setting standards for human embryonic stem cells. Science 2003; 300:913–916.
Jagerbauer EM, Fraser A, Herbst EW, Kothary R, Fundele R . Parthenogenetic stem cells in postnatal mouse chimeras. Development 1992; 116:95–102.
Hikichi T, Wakayama S, Mizutani E, et al. Differentiation potential of parthenogenetic embryonic stem cells is improved by nuclear transfer. Stem Cells 2007; 25:46–53.
Cattanach BM, Barr JA, Evans EP, et al. A candidate mouse model for Prader-Willi syndrome which shows an absence of Snrpn expression. Nat Genet 1992; 2:270–274.
Glenn CC, Porter KA, Jong MTC, Nicholls RD, Driscoll DJ . Functional imprinting and epigenetic modification of the human SNRPN gene. Hum Mol Genet 1993; 2:2001–2005.
Sakatani T, Kaneda A, Iacobuzio-Donahue CA, et al. Loss if imprinting of Igf2 Alters Intestinal Maturation and Tumorigenesis in Mice. Science 2005; 307:1976–1978.
Cassidy SB, Dykens E, Williams CA . Prader-Willi and Angelman syndromes: sister imprinted disorders. Am J Med Genet 2000; 97:136–146.
Matouk IJ, DeGroot N, Mezan S, et al. The H19 non-coding RNA is essential for human tumor growth. PLoS ONE 2007; 2:e845.doi: 10.1371/journal.pone.0000845.
Surani MA, Barton SC . Development of gynogenetic eggs in the mouse: Implications for parthnogenetic embryos. Science 1983; 222:1034–1036.
Allen ND, Logan K, Lally G, Drage DJ, Norris ML, Keverne EB . Distribution of parthenogenetic cells in the mouse brain and their influence on brain development and behavior. Proc Natl Acad Sci U S A. 1995; 92:10782–10786.
Kono T, Obata Y, Yoshimzu T, Nakahara T, Carroll J . Epigenetic modifications during oocyte growth correlates with extended parthenogenetic development in the mouse. Nat Genet 1996; 13:91–94.
Kono T, Sotomaru Y, Katsuzawa Y, Dandolo L . Mouse parthenogenetic embryos with monoallelic H19 expression can develop to day 17.5 of gestation. Dev Biol 2002; 243:294–300.
Kono T, Obata Y, Wu Q, et al. Birth of parthenogenetic mice that can develop to adulthood. Nature 2004; 428:860–864.
Hammond HA, Jin L, Zhong Y, Caskey CT, Chakraborty R . Evaluation of 13 short tandem repeat loci for use in personal identification applications. Am J Hum Genet 1994; 55:175–189.
Smith RN . Accurate size comparison of short tandem repeat alleles amplified by PCR. Biotechniques. 1995; 18:122–128.
Hwang WS, Ryu YJ, Park JH, et al. Evidence of a pluripotent human embryonic stem cell line derived from a cloned blastocyst. Science 2004; 303:1669–1674.
Stevens LC, Vamum DS, Eicher EM . Viable chimaeras produced from normal and parthenogenetic mouse embryos. Nature 1977; 269:515–517.
Fundele R, Norris ML, Barton SC, Reik W, Surani MA . Systematic elimination of parthenogenetic cells in mouse chimeras. Development 1989; 106:29–35.
Acknowledgements
This study was supported by the Hi-Tech Research and Development Program of China (863 Program 2006AA02A101).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Mai, Q., Yu, Y., Li, T. et al. Derivation of human embryonic stem cell lines from parthenogenetic blastocysts. Cell Res 17, 1008–1019 (2007). https://doi.org/10.1038/cr.2007.102
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2007.102
Keywords
This article is cited by
-
Significant differences in efficiency between two commonly used ionophore solutions for assisted oocyte activation (AOA): a prospective comparison of ionomycin and A23187
Journal of Assisted Reproduction and Genetics (2023)
-
Generation of developmentally competent oocytes and fertile mice from parthenogenetic embryonic stem cells
Protein & Cell (2021)
-
Generation of human androgenetic induced pluripotent stem cells
Scientific Reports (2020)
-
Identification and propagation of haploid human pluripotent stem cells
Nature Protocols (2016)
-
Pluripotent stem cells in disease modelling and drug discovery
Nature Reviews Molecular Cell Biology (2016)