Abstract
The effects of human EGFR to the malignant phenotype of human breast cancer cell line MDA-MB-231 were investigated experimentally. A retroviral vector containing a 5'1350bp fragment of the human EGFR cDNA in the antisense orientation was transfected into targeted cells by lipofectamine. The effects on cell proliferation, cell cycle and adherent ability to extracellular matrix (ECM) components were studied after the expression of antisense transcripts to EGFR 5'1350bp fragment in target cells. In vitro studies showed that the growth ability of the transfected cells was partialy inhibited in comparison to parental cells and to cells transfected with the plasmid containing the neomycin resistance gene only. It was found that EGF (10ng/ml) had an augmenation effect on the growth of transfected MDA-AS10 cells but not MDA-MB-231 cells. Flow cytometric analysis showed that the cell cycle of the transfected cells was abnormal with a decrease of cells in G2/M and S phases and an increase of cells in G1 phase, indicating a blockage in phase G1. Immunofluorescence of EGFR expression in transfectants stained with an anti-EGFR antibody was decreased and their growth in soft agarose was also severely impaired. The transfected cells showed less adherence to laminin (LN) and fibronectin (FN). In short, EGFR antisense RNA decreases the expression of EGFR on MDA-MB-231 cells and partially reverses their malignant phenotype as well.
Similar content being viewed by others
Introduction
Epidermal growth factor is a potent mitogen controlling the growth of various types of cells through their specific receptor1. The epidermal growth factor receptor (EGFR) has an intrinsic tyrosine kinase activity and is able to, upon ligand activation, phosphorylate itself and cellular substrates on tyrosine2. In addition to serving a normal physiological role, the EGFR has been consistently linked to cell transformation, both in experimental models and in human tumors. The EGFR gene is usually amplified and overexpressed in human tumors of ectodermal origin, suggesting an important role of EGFR in the development of these neoplasia3. Furthermore, EGFR overexpression is positively related with poor prognosis and high metastasis in human breast cancer4. This made EGFR a suitable target of tumor gene therapy. Studies of EGFR on human colon cancer cells, human rhabdomyosarcoma cells, human transfected KB cells and human hepatocarinoma cells have been reported5, 6, 7, 8. To directly investigate the role of EGFR overexpression in human breast tumor, we tried to decrease expression of EGFR in human breast cancer MDA-MB-231 cells. This paper reports that expression of antisense EGFR RNA results in a suppression of the transformed phenotype of MDA-MB-231 cells and the degree of suppression is positively carrelated to the decrease in expression of EGFR. In addition, expression of EGFR antisense RNA enables EGF to stimulate the growth of transfected MDA-AS10 cells.
Materials and Methods
Materials and cell culture
The human breast cancer MDA-MB-231 cells were maintained in 25 cm2 culture flasks at 37C in a humidified atmosphere of 5% CO2 in RPMI1640 medium supplemented with 15% new born bovine serum. Retroviral packaging cell line PA317 was cultured in DMEM medium supplemented with 15% new born calf serum.
Construction of antisense EGFR expression vector
Cleaved from pXEGFR (XbaI, HindIII), 4kb EGFR cDNA was ligated into XbaI and HindIII cloning sites of the vector pUC18 and designated as pUC18-EGFR. A 1350bp EGFR cDNA fragment was cleaved from pUC18-EGFR (BamHI) and ligated to retrovirus vector pLXSN. The recombined vector pLXSN-AE5' was used to transform E.coli DH5α, which were subsequently plated onto ampicillin-containing agar plates. Resistant colonies were selected and then expanded for plasmid DNA minipreparations. The orientation of the inserts was determined by restriction mapping.
Antisense transfection
Exponentially growing cells were washed with serum-free medium. The mixture of 200 μ1 medium containing 10 μl of lipofectamine (2 mg/ml GIBCO-BRL) and 4 μg of antisense EGFR expression vector DNA were added to the cells. Eight hours later, equal volume of culture medium was supplemented to cells. Twenty hours later, the cells were washed with serum-free medium and fed with fresh medium containing 15% new born calf serum. Forty-eight hours later, 400 μg/ml G418 was added to the medium for selection. After 2 ~ 3 wk, the G418 resistant clones were propagated in tissue culture. Antisense expressing vector pLXSN-AE5' transfected cells were designated as MDA-AS8 and MDA-AS10. Retrovirus expression vector pLXSN transfected cells were designated as MDA-pL.
PCR reaction
PCR was performed on genomic DNA extracted from parental and transfected cells. DNA concentration was determined by means of A260, and 0.5 μg was used for each PCR reaction in 50 μl final volume. The following primers were used to detect 433bp neo gene fragment: direct 5'-TCC ATC ATG GCT GAT GCA ATG CGG C-3' and reverse 5' -GAT AGA AGG CGA TGC GCT GCG AAT CG-3'. PCR reaction was performed with a 56 annealing temperature and 30 amplification cycles.
RNA isolation and reverse transcription-PCR
Total RNA was isolated from control cells and antisense clone cells according to method in " Molecular Cloning" 9. RNA concentration was determined by means of A260, and 5μg RNA were reversely transcribed using 200 ng reverse primers, 30 units of AMV reverse transcriptase, 20 units Rnasin (Promega) and 2.5 mM dNTP in a volume of 20 μl. The mixture was incubated at 42 water bath for 1 h, 2 μl reverse transcription products were used for following PCR reaction with the two primers: direct 5'-TTT CTG GCA GTT CTC CTC TC-3' and reverse 5'-CGC AGT TGG GCA CTT T-3'.
Cell proliferation assay
The cell count method described previously10 was used to examine the proliferation ability.
Flow cytometric study
Cells from subconfluent culture were resuspended, washed with PBS supplemented with 1% BSA (PBS-BSA), and incubated for 30 min on ice with a 1:40 dilution of anti-EGFR monoclonal antibody clone 528 (Oncogene Science, Uniondale, NY) in PBS-BSA. After washing with cold PBS-BSA, cells were incubated for 30 min on ice with a 1:50 dilution of FITC-conjugated goat antimouse immunoglobulin antiserum (Bonding, Beijing). Cells were again washed and resuspended in lml PBS, followed by analysis with a FACScan flow cytometer (Becton Dickinson). Each data shown was from 3 parallel experiments.
Growth in soft agarose
To examine anchorage-independent growth, 5 × 103 (MDA-MB-231, MDA-pLSXN, MDA-AS8 and MDA-AS10) cells were plated in triplicate in 24 well culture plates with 1 ml of culture medium, supplemented with 0.35% agarose. The cells in the dishes were incubated for 21 days at 37and then colonies were counted under microscope.
Cell cycle analysis
Cells from subconfluent culture were resuspended, washed with PBS and fixed in 70% ethanol for 30 min. Cells were washed, resuspended in PBS with RNase A (100μg/ml) and incubated in
30 for 30 min. Five microlitre propidium iodide (50 μg/ml, Sigma) were added to 106 cells for 20 min at 4 without light. The total DNA content per cell was assessed by analysis of fluorescence using a FACScan flow cytometer (Becton Dickinson). Effects of antisense EGFR on human breast cancer MDA-MB-231 cells
Adherence to different extracellular matrix proteins
Cells (2× 105/well) were seeded on 24-well plates precoated with either laminin (10 mg/ml in PBS) or fibronectin (10 mg/ml in PBS) for 1 h at 37 and 1% BSA for 2 h at room temperature. After 20 min, 40 min and 1 h incubation respectively at 37, the number of attached and unattached cells were counted.
Results
Construction of the expression vector
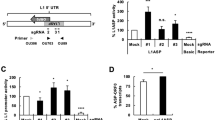
The sketch map of EGFR 5' 1350bp fragment inserted into pLXSN in antisense orientation was shown in Fig 1. Digested by EcoRI, the sense recombined plasmid was cleaved into 495bp and 6729bp fragments; the antisense recombined plasmid was cleaved into 887bp and 6337bp fragments (Fig 2).
Sketch map of EGFR 5' 1350bp cDNA inserted into retroviral vector pLXSN in antisense orientation.Amp and neo are selectable markers of vector in bacterium and mammalian cells respectively. The 1350bp fragment was inserted into BamHI site in antisense orientation.ψ+ indicates the extended retroviral packaging signal.
Integration and expression of antisense fragment
The presence of neo gene fragment in genomic DNA of different antisense transfectants but not parental MDA-MB-231 cells was shown by PCR (Fig 3). The presence of antisense transcripts in cellular RNA of MDA-AS8 and MDA-AS10 cells was shown by RT-PCR (Fig 4).
Effect of antisense EGFR RNA expression on the expression of cell surface EGFR
Cytofluorimetric analysis of membrane EGFR (Tab 1) showed a decreased expression in AS transfectants when compared to MDA-MB-231 or MDA-pL cells.
Effect of antisense EGFR RNA expression on cell proliferation
The proliferation ability of AS transfectants (Fig 5) was severely impaired. Plasmid pLXSN transfected cells had a similar growth rate comparable to that of the parental MDA-MB-231 cells. The proliferation ability was positively related to the expression level of membrane EGFR.
Growth effect of EGF on MDA-AS10 cells
MDA-MB-231 is a breast epithelial cell line which possesses a large amount of epidermal growth factor (EGF) receptor on the cell surface but does not respond to EGF. MDA-AS10 is an antisense cell clone transfected with recombined plasmid pLXSN-AE5'. The proliferation of MDA-AS10 was greatly reduced as shown in Fig 5. When cultured with EGF (10 ng/ml), MDA-MB-231 cells had the same proliferative rate as before, but proliferation of MDA-AS10 cells was enhanced rapidly as compared to that without EGF (Fig 6).
Cell cycle analysis
After stable expression of antisense EGFR RNA, the proportion of cells in the Effects of antisense EGFR on human breast cancer MDA-MB-231 cells
G0/G1 phase increased, and concurrently, the proportion of cells in the S phase decreased. It indicated that the cell cycle was blocked in G1 phase (Tab 2).
Effect of antisense EGFR RNA expression on growth in soft agarose
Compared with parental MDA-MB-231 cells and neo control MDA-pL cells, antisense EGFR RNA expression inhibited the growth of transfectants in soft agarose (Tab 3).
Ability of adherence to LN and FN was inhibited
At different time point, after the stable expression of antisense EGFR RNA, the ability of adherence of transfectants to LN and FN was respectively reduced, especially in the case of MDA-AS10 cells (Fig 7 and 8).
Discussion
EGFR has been implicated in the malignant behavior of many tumor types and its overexpression is positively related with poor prognosis and high metastasis in human breast cancer. Antisense technology is an important method to study the function of a specific gene11. Overexpression of a gene can be specifically inhibited by antisense RNA followed by studies on phenotype changes.
This study demonstrated that it is possible to downregulate the expression of EGFR in human breast cancer cells and decrease the proliferation ability of MDA-MB-231 cells by expression of antisense EGFR RNA. Moreover, it could be seen that the proliferation rate of different clone cells is positively related to the expression of EGFR on cell surface. Determined by Flow Cytometric studies, we found that in clone such as AS10 cells which possess low number of EGFR, the proliferation ability was more inhibited. This suggests that EGFR may play an important role in malignant proliferation of MDA-MB-231 cells.
It is generally acknowledged that the uncontrolled proliferation of tumor cells is related to the enhanced ability to synthesize DNA. Cell cycle analysis indicated that the two cell clones transfected with antisense EGFR had more cells in G0/G1 phases and less cells in S + G2/M phases, a result which implies that the malignant proliferation of transfected MDA-MB-231 cells were partially inhibited at G1phase.
MDA-MB-231 cells do not response to EGF (10 ng/ml) thought it has high-level EGFR12. However, in our studies, after being transfected with antisense EGFR, MDA-AS10 cells which exhibited low level of EGFR expression were able to response to EGF (10ng/ml). In human breast cancer cell line ZR75-1, Valverius et al found that there appears to be a threshold level of EGFR expression for the alteration in EGF responsiveness13. In addition, Davidson et al reported a tendency for EGF responsiveness only in those lines with fewer than 7 × 104 EGFR per cell, whereas cell lines with higher receptor numbers were generally unresponsive14. The mechanism of the correlation between EGF responsiveness and level of EGFR expression remains to be solved.
The invasion and metastasis of tumor cells is a complex process. Tumor cell invasion has been considered as a three-step process involving tumor cell adhesion, release of extracellular matrix (ECM) degrading enzymes and tumor cell motility15. Adhesion of tumor cells to ECM is an important step in the process of invasion. LN and FN are noncollagenous accessory glycoproteins of extracellular matrixes. It has been reported that adhesion of tumor cells to LN and FN had important influence on tumor metastasis16. It is interesting to note that after the stable expression of EGFR antisense RNA and the down-expression of EGFR on cell surface, the adhesion of transfected cells to LN and FN had been decreased respectively. This suggests that EGFR on MDA-MB-231 cells may influence cell invasion and tumor metastasis as well.
References
Gill GN, Bertics PJ, Santon JB . Epidermal growth factor and its receptor. Molecular and Cellular Endocrinology 1987; 51:169–86.
Koenders P, Beex L, Kienhuis C . et al. Epidermal growth factor and receptor and prognosis in human breast cancer: a prospective study. Breast Cancer Res Treat 1992; 23:134.
Huang S, Lin PF, Fan D, et al. Growth modulation by EGF in human colonic carcinoma cells: constitutive expression of the human EGF gene. J Cell Physiol 1991; 148:220–7.
Delaru JC, Friedman S, Mouriesse H, et al. EGFR in breast cancers: correlation with estrogen and progesterone receptors. Breast Cancer Res Treat 1988; 11:173–8.
Chakrabarty S, Rajagopal S, Shuang Huang . Expression of antisense epidermal growth factor receptor RNA down modulates the malignant behavior of human colon cancer cells. Clin Exp Metastasis 1995; 13:191–5.
Giovanni CD, Landuzzi L, Frabetti F, et al. Antisense EGFR transfection impairs the proliferative ability of human rhabdomyosarcoma cells. Cancer Res 1996; 56:3898–3901.
Moroni MC, Willingham MC, Beguino L . EGF-R antisense RNA blocks expression of the epidermal growth factor receptor and suppresses the transforming phenotype of a human carcinoma cell line. J Biol Chem 1992;267(5):2714–22.
Xu YH, Jiang WL, Peng SF, Cheng YH . Antisense EGF receptor sequence reverses the growth properties of human liver carcinoma cell line. Cell Research 1993; 3:75–83.
Sambrook J, Fritsch EF, Maniatis T . Molecular Cloning: A Laboratory Manual. 2nd ed. New York: Cold Spring Harbor Laboratory Press. 1989.
Zh E, Chen QG, Xu XL, et al. Tissue Culture Technology. 2nd ed. Beijing: People Hygiene Press. 1993.
Kopper L, and Kovalszkg I . Antisense tumor therapy. In Vivo 1994; 8:781–6.
Muller H, Loop P, Liu R, et al. Differential signal transduction of epidermal-growth-factor receptor in hormone-dependent and hormone-independent human breast cancer cells. Eur J Biochem 1994; 221:631–7.
Valverius EM, Velu T, Shankar N, et al. Over-expression of the epidermal growth factor receptor in human breast cancer cells fails to induce an estrogen-independent phenotype. Int J Cancer 1990; 46:712–18.
Davidson NE, Gelmann EP, Lippman ME, et al. Epidermal growth factor receptor gene expression in estrogen receptor-positive and -negative human breast cancer cell lines. Mol Endocrinol 1987; 1:216–23.
Liotta LA . Tumor invasion and metastases role of extracellular matrix. Cancer Res 1986; 46:1–7.
Akiyama SK, Olden K, Uamade KM . Fibronectin and integrins in invasion and metastasis. Cancer Metastasis Rev 1995; 14:173–89.
Acknowledgements
The project was supported by National Natural Science Foundation of China (No. 39470775)
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Fan, W., Lu, Y., Deng, F. et al. EGFR antisense RNA blocks expression of the epidermal growth factor receptor and partially reverse the malignant phenotype of human breast cancer MDA-MB-231 cells. Cell Res 8, 63–71 (1998). https://doi.org/10.1038/cr.1998.7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.1998.7