Abstract
The strategy of isolating the band-specific expression fragments from a probe pool generated by human chromosome microdissection was reported. A chromosome 14q24.3 band-specific single copy DNA pool was constructed based on this probe pool. Using total DNA of the pool as probe to hybridize the human marrow cDNA library, 68 primary positive clones were selected from 5 × 105 cDNA clones. Among these primary clones, 32 secondary clones were obtained after second-round screening and designed as cFD14-1 ˜ 32. Finally, 24 band-specific expression fragments were identified from these 32 positive clones by DNA hybridization. Those band-specific clones can hybridize to both 14q24.3 DNA and human genomic DNA but cann't hybridize to 17q11 ˜ 12 DNA. Partial sequences of 13 fragments of them were sequenced and identified as novel cDNA sequences, and these sequences were proved to have some homology with known genes in NCBI database. Analysis of expression spectrum of cFD14-1 suggested that the cDNA fragments thus obtained should be used to isolate the genes can not been cloned in 14q24.3 region.
Similar content being viewed by others
Introduction
The procedure of human gene cloning and identification has been greatly propelled by the rapid progress of human genome mapping and sequencing1. Positional cloning is the main strategy used in human gene cloning ,especially in disease-related gene cloning . Isolating genes from a disease-related chromosome region obtained by microdissection is an efficient way for positional cloning2, 3.
Chromosome 14q24.3 is an important region for many disease-related genes and cytokine genes such as c-fos, familial early-onset Alzheimer's disease related gene, palmoplantar keratoderma related gene and arrhythmogenic right ventricular cardiomyopathy gene (ARVDD) having been mapped in this area. Present paper reported 24 14q24.3 band-specific cDNAs were selected from a human fetal marrow cDNA library. These cDNAs can be used as the starting materials to clone 14q24.3 band-related genes furthermore.
Materials and Methods
Band-specific probe pools and primers
The human chromosome 14q24.3 probe pool and 17q11-12 probe pool were constructed separately by State Key Laboratory of Medical Genetics at Human Medical University. X-W primer (5'-CGG GAA TTC CTC TGC GAC ATG-3') was designed according to the probe pool constructing strategy. λgt11 primers are ISA-L 5'-GAC TCC TGG AGC CCG-3' and ISA-R 5'-CGC GGC CAG CGA TGG-3'.
Construction of single-copy DNA pool of 14q24.3
The secondary amplification and cloning of microdissected probe pool were referred to our previous work4. Filters containing latticed recombinants were hybridized to human genomic DNA. Positive clones being regarded as repetitive sequences were excluded. The second subtraction of repetitive sequences in negative clones collected after the first screening was performed as described above. The inserts of the negative clones selected after the second screening were cut out with EcoR I. Then the inserts were amplified with probe pool PCR system. A band-specific single-copy DNA pool was thus constructed.
Labelling of single-copy DNA pool
PCR labelling system was composed as follows: 0.4 μl (2U)of Taq polymerase, 1 μ1 of 25 μM X-W Primer, 5 μl of α-32P-dCTP (50 μCi), 1.5 μl of 2 mM dATP, dGTP, dTTP respectively, 5 μl of 10 × buffer, and 1 μl of band-specific single-copy DNA pool, which were made into total volume of 50 μl with sterile water. The mixtures were covered with 35 μl of mineral oil. PCR with 27 cycles (93 °C 1 min, 60 °C 30 sec, 72 °C 1 min) were performed, and a final extension of 72 °C 5 min. Four same PCR systems were performed.
Blocking of repetitive sequences in labelling probe
The labelled probe purified by Sephadex G-50 (with 500μl total volume) was mixed with 2 μl of human placenta DNA (25 mg/m1), 50μl of 5 M NaCl and 1.2 ml of precooled absolute alcohol. The mixture was precipitated at -80 °C for 15 min and centrifuged at 15,000 rpm for 10 min. After having been washed with 70% alcohol, the dried precipitant was resolved in 30 μl of TE (pH 8.0). After having been denatured in boiling water bath for 10 min, the DNA was renatured at 55 °C for 1 h and stored at 4 °C.
First and second screening of expression sequences
First screening was referred to Lambda Library Protocol Handbook of Clontech Corporation. Ten plates (diameter 150 mm) were spreaded with 50,000 phage plaques on each plate. Double filters were prepared. Prehybridization and hybridization were performed in formamide system5 at 37 °C. The filters were washed and autoradiography was performed. The positive plaque regions were transferred to 1000 μl of SM solution with 20 μl of Trichloromethane and stored at 4 °C.
Plates (diameter 90 mm) were spreaded with 500 plaques on each plate according to the titers of the first screening clones. Double filters were prepared. Prehybridization and hybridization were the same as the first screening procedure. According to the autoradiography results, mono-clone plaques were transferred to 200 μl of SM solution containing 4 μl of Trichloromethane and stored at 4 °C.
Identification of positive clones by DNA hybridization
Probes were prepared by amplification of phage inserts. The reaction system contained 0.4 μl (2U) of Taq polymerase, 1 μl of 25 μ M λ gt11 primer, 2 μl of 5 mM dNTP, 5 μl of 10×buffer, and 1 μl of phage solution, which were made into total volum of 50 μl with sterile water. The system was covered with 35 μl of mineral oil. PCR with 35 cycles (93°C 1 min , 55°C 1 min, 72°C 2 min) were performed and a final extension of 72°C 5 min.
Dot blot membranes loading 14q24.3 DNA, 17q11-12 DNA and human genomic total DNA (HGT DNA) were hybridized with human genomic DNA probe first. Then, after having been washed, these membranes were hybridized with isolated cDNA probes respectively. Eight membranes were prepared and each was hybridized with 4 cDNA probes in turn.
Sequencing and homology comparison of partial cDNAs
cDNA fragments identified by DNA hybridization were cloned into pUC119 vector and sequenced. Homology comparisons were performed with NCBI (National Center of Biology Information) data base.
Northern blot
cFD14-1, which was confirmed to be a 14q24.3-specific expression sequence by DNA hybridization, was hybridized to Multiple Tissue Northern (MTN) membrane as follows: MTN membrane was prehybridized in prehybridization solution (5 × SSPE, 10 × Denhardt's solution, 50% formamide, 2% SDS, and 100 μg/ml CT DNA)at 42 °C for 4 h. Then the prehybridization solution was replaced by hybridization solution (5 × SSPE, 10 × Denhardt's solution, 50% formamide, 2% SDS) containing labelled probe and the hybridization was performed at 42 °C for 20 h. The membrane was washed at following condition: Washing twice with 2 × SSC/0.05% SDS at 42 °C for 15 min each, then twice with 0.1 × SSC/0.1% SDS at 50 °C for 30 min. Autoradiography was performed.
Results
Construction of human chromosome 14q24.3 single-copy DNA pool
The secondary amplified product of 14q24.3 probe pool was presented as "smear" in electrophoretic agarose gel with the size ranging from 300bp to 650bp. The amplified product was cloned into pUC119 and the recombinant clones were arranged in lattice on nylon membranes. Colony hybridization was performed with human genomic DNA probe. Results were shown in Tab 1.
Inserts of negative clones selected after two rounds repetitive sequences screening were cut out with EcoR I.Then single-copy DNA pool was constructed by amplifing the recovered inserts.
Screening of expression sequences
Human marrow cDNA library was screened using labelled 14q24.3 single-copy DNA pool as probe. Sixty-eight primary positive clones were selected after the first screening and 32 secondary positive clones were obtained after the second screening.
Identification of secondary positive clones by DNA hybridization
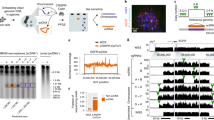
To identify the 14q24.3 band-specificity of the 32 secondary positive clones, DNA hybridizations were performed. (Fig 1, Tab 2). And, 24 clones hybridizing to 14q24.3 DNA and human total genomic DNA showed strong hybridization signals but no signal was shown to 17q11-12 DNA. Therefore, these clones were confirmed to be the single-copy sequences in the band 14q24.3.
Hybridized identification of candidate clones of human chromosome 14q24.3 band specific expression sequences. A. B.C.D are dot-blot membranes carried with the same DNA samples. These samples are 1: 14q24.3 DNA, 2: 17q11-12 DNA, 3: Human Genomic Total (HGT) DNA. Each membrane was hybridized with HGT and 4 cFD14 probes, the DNAs with hybridized signals only on sample 1 and 3 were regarded as 14q24.3 specific sequences, while on sample 1, 2 and 3 as repetitive sequences.
Sequencing and homology comparison of partial cDNA fragments
Thirteen cDNAs were partially sequenced and homology comparison of these sequences were perfo rmed with NCBI database. Results are shown in Tab 3.
Northern blot of cFD14-1
The expression pattern of cFD14-1 is shown in Fig 2. The size of transcripts in 8 tissues has some differences. For an example, 4 transcripts with different size are shown in kedney while only 2 transcripts (1.0kb and 2.4 kb )in heart.
Discussion
Subtraction of repetitive sequences from chromosome band probe pool
Human genome contains about 35% ˜ 40% repetitive sequences6 and so does microdissected chromosome band. In cDNA library there is also about 3% ˜ 5% repetitive sequences7. If hybridized signals of repetitive sequences can not be blocked efficiently, it is difficult to isolate cDNA from cDNA library. So the keypoint of isolating expression sequences from a band specific probe pool is to get rid of the interference of the repetitive sequences.
Improvements in two facets are reported here. The first is that construction of a single-copy DNA pool can remove most of the repetitive sequences in microdissected band. The second is to block low copy-number repetitive sequences with human placenta DNA in a small reaction volume8.
The advantages of microdissected probe pool in cloning disease-related genes
There are about 50,000 ˜ 100,000 genes in a human genome6. Using microdissected chromsome probe pool, band-specific cDNAs which might be the candidate of disease-related genes can be batchly isolated. In addition, since there are many gaps in the YAC-contigs map of human genome9, microdissected chromosome probe pool is useful for cloning the genes located in the gaps.
For there are several important functional genes mapped in chromosome 14q24.3 (GDB database), we chose this band as the target area to perform positional cloning of genes. The microdissected human chromosome 14q24.3 probe pool was afforded by State Key Laboratory of Medical Genetics at Hunan Medical University. The probe pool was proved to be 14q24.3 precise by flouorescence in situ hybridization (FISH) (Data not shown). And 24 band-specific cDNAs were isolated. After sequencing and homology comparison of these cDNA clones, 13 of 24 band-specific cDNAs were proved to have some homology with known genes (Tab 3). For example, cFD14-5 has had 8l % homology with human CALM1 gene10 in 110bp, the homology between cFD14-6 and human estrogen receptor protein related gene11 reaches 93 % in 231bp and cFD14-10 has 82 % homology with human BNSP gene12 in 132bp. All these known genes have been mapped on other chromosomes, which infers that the cDNA fragments reported here may represent the gene having the similar structure or function as those genes. But in these cDNA clones, no one is the same to these known genes. The proper reason is the 14q24.3 band is too large (about 12.8 Mb) and the number of known genes located in this band is small (GDB database). In addition, we have recently isolated a novel STR marker D14S1435 from this microdissected probe pool in another project13 and relocalized this STR on 14q24.3 accurated by FISH. This result further confirmed the good quality of the probe pool and the band-specific nature of cDNAs. Furthermore, the expression patterns of cFD14-1 showed this fragment was differentially expressed in many tissues (i.e. the hybridizing signal in heart is much stronger than that in the other tissues and the signal in brain is too weak to detect) and the phenomenon of RNA alternative splicing or homologous genes might be existed. All these suggest that the cDNAs obtained in this way should be helpful to isolate uncloned genes to be mapped in the specific chromosome 14q24.3 band.
References
Casey D . HGMIS, and NCHGR and DOE OHER program staff Five Years of Progress in the Human Genome Project. Human Genome News 1995; 7:4–9.
Deng HX, Yoshiura KI, Dirks RW, Harada N, Hirota T, Tsukamoto K, Jinno Y and Niikawa N . Chromosome-band specific painting: chromosome in situ suppression hybridization using PCR products from a microdissected chromosome band as a probe pool. Hum Gene 1992; 89:13–7.
Kao FT, JW Yu . Chromosome microdissection and cloning in human genome and genetic disease analysis. Proc Natl Acad Sci USA 1991; 88:1844–8.
Deng Y, Yu L, Wang B, Yuan HY, Han SS, Qiao YH, Zhao SY . Batch isolation of single- copy DNA fragments by using PCR-SSCP from the probe pool of human chromosome 17q11-12 generated by microdissection. Journal of Fudan University (Natural Science), 1995; 34:370–8.
Yu L, Wang WY, Wang QB, Wang MQ, Yang YM, Zhu JD, Zhao SY . The preliminary study on RFLPs of Chinese Dystrophin gene. Acta Biologiae Experimentalis Sinica, 1992; 25:173–80.
Zhao SY . Human genome. Shanghai Science and Technology Publishing House. 1993: 1–41.
Crampton J, Davies K, Knapp T . The occurrence of families of repetitive sequences in a library of cloned cDNA from human lymphocytes. Nucleic Acid Res 1981; 9:3821–6.
Zhang M, Yu L, Hu PR, Shi SL, Wu GJ, Zhao SY . An improved method of preparation of large genomic DNA probe and its application. Chin J Med Genet 1997; 14:112–4.
Chamakov IM, P Rigault, IL Gall, et al. A YAC contig map of the human genome. Nature 1995; 377s:175–298.
Rhvner JA, Ottiaer M, Wicki R, Greenwood TM and Dtrehler EE . Structure of the human CALMI calmodulin gene and identification of two CALM 1-related pseudognenes CALM1P1 and CALM1P2. Eur J Biochem 1994; 225:71–82.
Dotzlaw H, Alkhalaf M, Murphy LC . Characterization of estrogen receptor variant mRNAs from human breast cancers. Mol Endocrinol, 1992; 6:772–785.
Kim RH, Shapiro HS, Wrana JJ, Sodek J . Characterization of the human bone sialoprotein (BNSP) gene and its promoter sequence. Matrix Biol 1994; 14:31–40.
Zhang M, Yu L, Hu PR, Zheng QP, Sun XY, Zhao SY . Isolation and polymorphic fre- quency detection of a novel STR on human chromosome 14q24.3. Chinese Science Bulletin 1997; 42:2328–31.
Acknowledgements
The project was supported by the National Natural Science Foundation of China (No: 39525015), the Foundation for Shanghai Technology Development Key Project (No: 779614001) and 863 High Technology Programe (No: 102100401).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Zhang, M., Yu, L., Hu, P. et al. Isolation of 24 novel cDNA fragments from microdissected human chromosome band. Cell Res 8, 135–142 (1998). https://doi.org/10.1038/cr.1998.13
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.1998.13