Abstract
Recently the antichromosome antisera from several scleroderma patients have been found to recognize the pellicle of metaphase and anaphase chromosomes. In order to identify the pellicle components, we used these antichromosome antisera to screen a human embryonic cDNA library. The sequences of the positive clones are identical to the cDNA gene sequence of CENP-C (centromere protein C), a human centromere autoantigen. This result suggusts that CENP-C is a component of the pellicle of human metaphase and anaphase chromosomes.
Similar content being viewed by others
Introduction
During normal cell division, the chromosome splits evenly, giving the same genetic material to each daughter cell. The accuracy of such cell division depends on the interaction between the spindle apparatus and the cis-acting region of each chromosome known as the centromere. Many essential mitotic functions occur at or are controlled by the centromere. Capturing spindle microtubules, the growth and disassembly of microtubules, the lining up of chromosomes on the metaphase plate, and the splitting off of the sister chromatids during mitosis are all somehow regulated by the centromere1. A physical structure known as the kinetochore, is found at the surface of the centromeric heterochromatin. The kinetochore consists of three layers which mediates attachment of the chromatin to the spindle microtubules2. One of the recently identified mammalian centromere proteins (CENPs) is centromere protein C (CENP-C), a component of the trilaminar kinetochore localized to the inner kinetochore plate3.
The chromosome pellicle is a peripherial structure of mammalian metaphase and anaphase chromosomes. This structure cannot be observed by conventional cytological methods, but it can be detected by immumofluorescent staining with the pellicle antisera obtained from some scleroderma patients4, 5, 6, 7. It was found that the composition of the pellicle is quite complex, consisting of many different proteins8. Recently, in the course of analyzing these proteins, we discovered that CENP-C, the main component of the kinetochore, is one of them. Pellicles exist on metaphase and anaphase chromosomes of mammalian cells. The antichromosomal antisera from scleroderma patients has been shown to recognize the pellicles of metaphase and anaphase chromosomes5. In order to identify which pellicle proteins are recognized by these antibodies, we screened a human embryonic cDNA expression library with four antichromosome antisera from the scleroderma patients. Five positive clones were obtained, and the sequence of each positive clone was identical to the human CENP-C cDNA gene sequence.
Materials and Methods
Cell culture
Mouse L929 cells and human Hela cells were cultured over coverslips with 5 % CO2 in Dulbecco's modified Eagle's medium supplemented with 20 % fetal calf serum, 100 μg/ml penicillin, 100 μg/ml streptomycin and 0.25 μg/ml fungizone.
Antibodies
Anti-chromosome sera were screened from scleroderma patients by indirect immuno-fluorescent staining. Among 338 patients, 12 have anti-chromosome sera. In this experiment, all 4 samples of the sera targeted the chromosomal pellicle8. The secondary antibody used was rabbit anti human IgG, conjugated with fluorescein isocyanate.
Embryonic tissue
Three month embryonic tissue was kindly received from the Department of Pathology in Shanghai International Peace Women and Children's Hospital. The tissue was verified by tissue slides.
Construction of human embryonic cDNA library
Human embryonic mRNA was isolated using an mRNA Purification kit (Pharmacia Biotech), and cDNA synthesized. The cDNA library was constructed using ZAP-cDNA synthesis Kit and Gigapack II Plus Packaging Extract (Strata-gene) according to the instructions provided with the kits.
Library screen using patient sera
Library screening was carried out using E.coli BB4, and 106 phage were screened with approximately 30,000 pfu used for each large 150 mm plate.
The inverted 150mm plates were incubated at 42 °C for 3.5 h or until pin-sized plaques were seen. Nitro-cellulose filters were prepared prior to incubation, by dipping in 10 mM IPTG and air dried. The filters were placed on agar. Holes were poked in the agar and filters for orientation. The plates were incubated at 37 °C for an additional 3.5 h. First, the filters were removed and washed in TBST. Duplicate filters were placed on the original agar and incubated for an additional 4 h. The filters were washed 3-5 times in TBST 10-15 min each time, and were put in blocking buffer (1% BSA TBST) for 1 h or overnight. The filters were then put in fresh blocking buffer with an appropriate antibody dilution (human sera 1:1000), and were shaken at room temperature for 1 h. The filters were washed again in TBST 3 times (10-15 min each time), and were then incubated with the secondary antibody (Alkaline Phosphatase conjugated or Horseradish Peroxidase conjugated Goat anti-human monoclonal antibody) diluted in blocking buffer (1:3000) and were shaken at room temperature for 1 h. Finally, the filters were washed with TBST 3 times (10-15 min each time), and developed9. Positives were picked out using a 1 ml pipet to core out the agar, and dropped in to 1 ml SM buffer and stored at 4 °C . Second and third screenings of positives were performed to confirm the single positive plagues.
Excising insert from lambda ZAP
20 ml of 2YT with 500 μl of XL-1 overnight culture was inoculated. It was shaken at 37 °C until OD600 = 1.0. In a 50 ml conical tube, 200 μ1 XL-1, 200 μl phage and 1 μl VCSM13 helper phage were mixed and incubated at 37 °C for 15 min. For control, 200 ml phage was replaced with 200 ml SM buffer (without phage). 5 ml 2YT was then added, and incubated 3 h 37 °C with shaking at 300 rpm. For excising the insert from Lambda ZAP, the tubes were incubated at 70 °C for 20 min and were centrifuged at 4000 g for 5 min. The supernatant obtained was mixed with XL-1 cells. a) 10 μl of phagemid with 200 μ1 XL-1; b) 20 μ1 of 10−2 phagemid with 200 μ1 XL-1; and was incubated at 37 °C for 15 min. 100 μ1 of the mixture was plated on LB/Amp plate and incubated overnight at 37 °C for the positive colonies. Standard Alkaline Lysis Procedure was used for minipreparations. The plasmid was doubly digested with KpnI and SacI respectively, and incubated at 37 °C for 1 h and 1% agarose gel was run to ensure that the positive clones were enclosed in the inserts.
Sequencing of inserts for identification
Double stranded sequencing was performed using T7 DNA polyrnerase on AutoRead (Pharma cia). The DNA sequences were sent to blast server on network for sequence comparison. All five positive clones were identified as CENP-C.
Indirect immunofluorescent staining
The coverslips with cells, after a short rinse in PBS at pH 7.0, were fixed with 3 % paraformaldehyde in the PBS for 5 min, followed by treatment with 1% Triton X-100 in the PBS for 20 min, and washed in the PBS for 3 times (4 min each) Then, the fixed and washed cells were incubated with primary antibody for 45 min at room temperature and washed with PBS for 3 times (4 min each). They were then incubated with secondary antibody for 45 min at room temperature. After 3 times (4 rain each) washed with PBS they were counterstained with Hoechst 33258 11.5 μg/ml for 2 min and mounted in glycerol PBS phenylene diamine.
Results
Cloning, CENP-C from human embryonic cDNA library
A human embryonic cDNA library was constructed, mRNA was isolated from three months human embryonic tissues. Tissue cDNA was synthesised. EcoR1 Adapter and Xho I Adapter were used for ligation and after packing the recombinant lambda l08 pfu were obtained. The length of the average insertion fragments was between 0.5 - 1.5kb.
Scleroderma patient sera were used to screen this human embryonic cDNA library. 5 positive clones were isolated from 106 plaque. These positives were all confirmed with secondary and tertiary screenings. The inserts were excised from Lambda ZAP and were currently in bluescript plamid. Minipreps of the phagemids were performed and doubly digested with Kpn I and Sac I, which were sited at the ends of the inserts in bluescript plasmids, yielded inserts of sizes ranging from approximately 1.6kb to 1.8kb. Minipreps of the clones were done, and sequencing of the DNA was done using Pharmacia Auto Read Sequencing. The sequences, an average of about 300bp in length, were sent to the blast server on the internet, and all the clones were determined to be parts of CENP-C.
Distribution of CENP-C on metaphases of human and mouse chromosomes
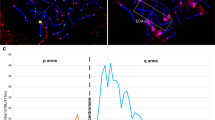
The same scleroderma antisera that was used to recognize the pellicle of the mouse metaphasic and anaphasic chromosomes screened in our previous investigation were used8 to screen a hnman cDNA library. To compare the antigen distribution of the mouse cells (L929) with that of the human cells (Hela), indirect imInunofluorescent staining was made on both mouse and human cells. The results are shown in the figures. It can be seen from Fig 1 that in the human interphase cells the antigens are predominatly found in the nuclei, forming a large number of speckles while apparently very little are found in the cytoplasm. However, in the mouse cells there are an abundant antigens in the cytoplasm, expressing as lumps and a lesser amount in the nuclei appearing as weakly stained particles. At metaphase and anaphase all the human chromosomes were decorated by the antiserum, as shown in Fig 2, while only the pellicle of mouse chromosomes were stained as reported previously8. The difference between human and mouse chromosomes in the distribution of the antigen may not be real. Unfortunately, no antiserum is available for further investigation.
Discussion
Although many years ago the pellicle structure of metaphasic and anaphasic chromosomes was proposed4. Its existence as chromosome structure has been definitively demonstrated only in recent years by indirect immunfluorescent staining with specific antisera from scleroderma and lupus erythromatosis patients5, 6, 7, 8. The proteins of this structure are quite complex. By immunobloting with the antisera specific to the pellicle from scleroderma patients, we identified 19 proteins with different molecular weights8. These proteins come from different cellular structures of interphasic cells, including cytoplasm, nucleolus and other nuclear structures5, 6, 7. However, none of them has been identified to date. The purpose of the present study is to attempt to characterize these proteins first by screening the cDNA from expressed cDNA library by the antipellicle antiserum identified in our previous investigation8. Then, their chemical compositions are deduced from the sequences of the cDNAs. In this way, we can also isolate their cDNA genes. At present, only one of the 19 proteins recognized by the antipellicle antisera was screened from an expressed human cDNA library. From the cDNA sequence, the protein was identifled as CENP-C, one of the kinetochoreprotiens with a molecular weight of 140 KD. This is one of the 19 proteins detected by immunobloting in our previous work8.
When the mouse metaphasic and anaphasic chromosomes were stained using one of the antipellicle antiserum, we failed to differentiate the centromeres from the other part of the pellicle8. It is known that CENP-C is a protein component of all mammalian kinetochore. Thus, the results obtained suggest that CENP-C is a protein component of the mouse chromosome pellicle. Furthermore, since CENP-C is situated at the inner layers of the kinetochore, the whole kinetochore proteins may also be the components of the pellicle. Unfortunately, the pellicle of human mataphasic and anaphasic chromosomes cannot be demonstrated clearly because of the lack of the antipellicle antisera.
References
Bloom K . The centromere frontier: kinetochore components, microtubule-based motility, and the CEN-value paradox. Cell 1993; 73:621–24.
Saitoh H, Tomkiel J, Cooke CA, Ratrie III H, Maurer M, Rothfield NF, Earnshaw WC . CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell 1992; 70:115–25.
Tomkiel J, Cooke CA, Saitoh H, Bernat RL, Earnshaw WC . CENP-C is required for maintaining proper kinetochore size and for a timely transition to anaphase. JCB 1994; 125:531–45.
Tsu TC, Arriglui FE, Klevecz RR et al. The nucleolus in mitotic divisions of mammalian cells in vitro. J Cell Biol 1965; 26:539–53.
Shi Luji ( L. C. Sze ), Ni Zumei, Zhao Shi et al. Involvement of a nucleolar component, perichromonucleolin, in the condensation and decondensation of chromosomes. PNAS 1987; 87:7953–6.
Ni Zumei, Je Little, Shi Luji ( L. C. Sze ). Assembly and disassembly of mammalian chromosome pellicle. Cell Research 1992; 2:195–201.
Ni Zumei, Su lide, Shi Luji ( L. C. Sze ) et al. The Localization of antisera of scleroderma patients and their antigen in cell structure. J of Chinese Medicine 1993; 73:270–3.
Wei Ying, Ni Zumei, Zhou Yan, Wu Li, Shi Luji ( L. C. Sze ). The study of metaphasic chromosome pellicle. Acta Biologiae Experimentalis Sinica 1994; 27:505–13.
Sambrook J, Fritsch EF, Maniatis T . Molecular Cloning: a Laboratory Manual. 2nd Edition, Cold Spring Harbor Laboratory Press, Cold Spring Habor, New York. 1989.
Acknowledgements
This research was supported by the State High Tech Development Program (863) Foandation of China and RGC Research Grant of Hong Kong.
Author information
Authors and Affiliations
Corresponding author
Additional information
*This article is dedicated to Dr. Lu Ji SHI (L. C. Sze) in honor of his 80th birthday.
Rights and permissions
About this article
Cite this article
Xie, Y., Ni, Z., Gu, J. et al. Human chromosome pellicle antibody recognizing centromere protein-C (CENP-C), the main component of the kinetochore. Cell Res 7, 13–19 (1997). https://doi.org/10.1038/cr.1997.2
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/cr.1997.2