Abstract
This article is one of the serial studies on the characteristics of the molecular structure for dystrophin gene in Chinese. By using the entire dystrophin cDNA (14 kb) as a probe, the number and RFLPs of Bgl II exon-containing fragments of the dystrophin gene were analysed. Four new Bgl II fragments were found, two of them (3.7 and 6.2 kb) detected by comparing the hybridization patterns with cDNA1–2a, la and 2a, one (9.3 kb) from the hybridization pattern with cDNA 9 by lengthening migrating distance of DNA fragments in electrophoresis , and another one (4.0 kb) by comparing the patterns with cDNA 11–14, 11a, 11b, 11c-12a and 14. The results indicated that the number of Bgl II exon-containing fragments should be 59 rather than 55 reported previously, which laid the foundation of the Bgl II partial restriction map for dystrophingene. Three of the four RFLPs found in Caucacian appear in the hybridization patterns of three subclones, i.e. cDNA 2b-3, cDNA 4–5, and cDNA 5b-7.The values of expected heterozygote frequency (EHF) were 0.33, 0.33 and 0.40, and the observed heterozygote frequency (OHF) were 0.40, 0.40 and 0.48 respectively. Meanwhile, two new rare allelic fragments (15 kb) were found in RFLPs from Bgl II/2b-3 and Bgl II/4–5a patterns respectively. These Bgl II RFLPs and four XbaI RFLPs documented in our laboratory have been used to detect the carrier in 7 DMD families and 1 BMD family. Of the 69 individuals from the 8 families, 11 females were diagnosed as the carriers with DMD mutation, 4 females as the doubtful carriers, 12 females were defined as normal genotype and 2 females as probably normal. The results suggest that the carrier testing method based on dosage intensity analysis and genotype analysis by using dystrophin cDNA as a probe will be more sensitive and accurate.
Similar content being viewed by others
Introduction
Duchenne and Becker muscular dystrophy (DMD, BMD) are X-linked recessive disorders, affecting about 1 in 3300 liveborn males1. The gene has been located at Xp21 by identification of unusual patients with cytogenetically visible deletion of Xp212 or balanced X: autosome translocation3. Subsequent linkage studies using randomly cloned DNA probes from this region confirmed the localization of the gene to Xp21 4, 5. Deletions of the DMD gene in 17% of individuals with Duchenne muscular dystrophy (DMD) were first detected with the single-copy DNA probes pERT876, 7, 8, XJ9, JBir and J66-H1 8. After the complete 14 kb cDNA was cloned10, gene deletions were detected in 60–65% DMD/BMD individuals 11, 12, 13, 14, 15, 16. Detecting the gene deletion with cDNA probes could also be applied in prenatal diagnosis and carrier identification at DNA level 17, 18. The accuracy of carrier testing based on deletion analysis by gene dosage comparison should achieve 100% theoretically, but actually the restriction-fragment pattern revealed by cDNA probes is quite complex because of the large size and numerous exons of the dystrophin gene. Deletion of the small co-migrating fragments is usually missed. Particularly, a significant number of DMD/BMD families (30%-40%) without detectable deletion or duplication should be investigated by RFLP linkage analysis also.
With entire 14 kb dystrophin cDNA12 as the probe, the RFLPs can be used in families with or without deletion / duplication for the carrier detection and prenatal diagnosis. Therefore, it is very important to construct the partial restriction maps with the restriction enzymes which can show RFLPs with high polymorphism information content (PIC) value at restriction sites within the gene. The hybridization pattern produced by eight cDNA probes, which cover the complete dystrophin gene, and eight restriction enzymes were analysed. The results demonstrated the usefulness of combining the genetic linkage analysis with dosage intensity for carrier testing12, 19. Based on Bgl II and Xba I RFLP patterns, the carrier status of 29 females in 1 BMD family and 7 DMD families was established. The present paper reported the number of Bgl II exon-containing fragments, the frequency of three Bgl II RFLPs and the result of DMD/BMD carrier testing in Chinese.
Materials and Methods
To find new restriction-fragment in Bgl II /cDNA hybridization pattern, 7.5μg DNA from each of 100 unrelated healthy individuals (male 60, female 40) and 69 individuals from one BMD family and seven DMD families was digested with Bgl II, or Xba I (Bio-labs) and hybridized with the dystrophin cDNA subclones 1–2a (including 1a, 2a subprobes), 2b-3, 4–5a, 5b-7, 8, 9, 10 and 11–14 (including 11a, 11b, 11c-12a, 14 subprobes) from American Type Culture Collection.
Genomic DNA was extracted from the whole blood leucocytes anticoagulated in EDTA21 and then transferred overnight to nitrocellulose filter (Amersham Inc.) by Southern blotting. Electrophoresis, hybridization, and filter washing were carried out according to methods described elsewhere22. Autoradiography with intensifying screens was exposed at −70°C for 3–7 days.
Subprobe preparation: subclone p1a (0.4 kb) and a 1.1 kb fragment were cut by BamHI from insert 1–2a; p2a (0.7 kb) subclone and a 0.8 kb fragment were cut by EcoR V from insert 1–2a; p9 (1.2 kb), p10 (0.9 kb) and p11–14 (4.2 kb) subprobes were cut by BamHI from cDNA 9–14; the p11 (1.37 kb) was cut by PvuII from cDNA 9–14. p11a (0.53 kb) and p11b (0.84 kb) subprobes were cut by PstI from the p11 fragment; the p11c-12a (0.4 kb) subprobe was cut by Bgl II from cDNA 9–14, and p14 (1.0 kb) was cut by Hind III from cDNA 9–14. The cDNA 1–2a and 9–14 were two of six clones which constituted the entire 14 kb dystrophin cDNA from ATCC. To determine the allelic frequency of Bgl II PFLPs in Chinese, those normal individuals mentioned above were tested. To verify the effect of combining linkage analysis with dosage intensity on carrier testing, 69 DNA samples from 8 DMD/BMD families digested with Bgl II and Xba I were analysed using dystrophin cDNA as the probe.
Results
1. Counting exon-containing Bgl II fragaments
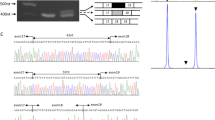
The hybridization pattern revealed by 14 subprobes were shown in Tab 1 and Fig 1. Four new Bgl II fragments were found. Among them, both 3.7 kb and 6.2 kb fragments were identified by comparing with the patterns from subprobe 1a, 2a and 1–2a; a 9.3 kb fragment was detected by lengthening the electrophoretic distance in Bgl II /9 pattern; and the 4.0 kb fragment was detected by comparing with the six patterns from subprobes 11a, 11b, 11c-12, 12a, 14 and 11–14.
Normal human Bgl II restriction patterns revealed by the fourteen adjacent segment of the dystrophin cDNA, the arrows point to various fragments arisen from Bgl II RFLPs, subprobe 1a and 2a were isolated from cDNA 1–2a. subprobe 9, 10 and 11–14 were isolated from cDNA 9–14, subprobe 11a, 11b, 11c-12a and 14 were isolated from cDNA 11–14 fragments.
The sum of all Bgl II fragments was fifty-nine in dystrophin gene not counting the five fragments which appeared in two boundary patterns at the same time. These five fragments are 5.0 kb fragment appeared in two patterns from cDNA 1–2a and cDNA 2b-3; 3.1 kb in patterns from cDNA 4–5a and cDNA 5b-7; 3.5 kb in patterns from cDNA 5b-7 and cDNA 8; 6.1 kb in patterns from cDNA 9 and cDNA 10 and 1.4 kb in patterns from cDNA 10 and cDNA1 1–14. In the hybridization pattern, the first band was 3.7 kb containing 33 bp cDNA, and the last band was 18.5 kb containing 2718 bp cDNA in 59 Bgl II exon-containing fragments of dystrophin gene.
2. Bgl II polymorphisms
Three of four Bgl II RFLPs found in Cancasian19 appeared in hybridization pattern of three subclones, i.e. cDNA 2b-3, cDNA 4–5, and cDNA 5b-7. The size and frequency of allelic fragments, expected heterozygote frequency (EHF) and ob- served heterozygote frequencies (OHF) were shown in Tab 2. Because EHF and OHF are of high PIC value, the three Bgl II RFLPs will be very useful in carrier testing and prenatal diagnosis. Furthermore, two rare new allelic fragments were found in both Bgl II RFLPs with cDNA 2b-3 and cDNA 4–5a, and shown in Fig 1, Fig 2 and Tab 2.
3. Carrier detection
Using the dystrophin cDNA and restriction enzymes Bgl II and Xba I, the genotype of 69 subjects from 1 BMD and 7 DMD faimlies, who asked to identify their carrier state for DMD /BMD gene mutant was investigated. According to family history, the 8 families were divided into types A, B and C. Type A was defined as patients appeared in two successive generations in a family. Type B involved at least 2 patients in one generation and type C is only a proband suffering from the disease in a family. 69 genotypes from the 8 families are showed in Fig 3–4. Four probands show partial deletion in dystrophin gene (Tab 3). Based on genotype and dosage intensity analysis, 29 females in the 8 families were diagnosed. The results of carrier testing were shown in Tab 3. Of the 29 subjects, 11 females were diagnosed as the carriers with DMD gene mutation, 4 females as the doubtful carriers, 12 females as normal, 2 females as probably normal. 3 boys less than 3-year old were diagnosed as normal individuals. The recombination events occuring in DMD gene were found in 2 females of 40 offsprings. A novel Xba I/ 1–2a polymorphism originated from paternal germ cell was found in 1 family.
Information map for eight Bgl II RFLPs of dystrophin gene in eight DMD/BMD families. “Aa” represents RFLP from XbaI/1–2a, “Bb” from Xba I/2b 3, “Cc” from Bgl II/2b-3, “Dd” from Bgl II/4–5a, “Ee” from Xba I/4–5a, “Ff” from Bgl II/5b-7, “Gg” from Xba I /10, “Hh” from Bgl II/11–14. “del” represents the deletion mutation. “A1” represents a BMD family, “A2–C1” represents seven DMD families. Black box means allelic fragments showing in hybridization pattern, open box represents no allelic fragments shown.
The pedigrees and genotypes of eight DMD / BMD families. The proband of A1 family suffered from Becker muscular dytrophy. The probands of A2, B1, B2, B3, B4, C1 and C2 families suffered from Duchenne muscular dystrophy, with black boxes representing patients with DMD or BMD and arrow pointing to the proband in the family; ring with black dot at the centre of circle representing diagnosed carrier with DMD /BMD mutation. In genotype analysis, A–H represented each of the eight RFLPs same as Fig 3. The capital letter means allele fragment with larger molecular weight in RFLPs, and the small letter means allele fragment with smaller molecular weight.
Fig 5 showed a part of the hybridization patterns from carrier testing. Bgl II /2b-3, Bgl II/ 4–5a, Xba I/ 1–2a patterns were derived from family A1 with BMD, Bgl II /5b-7 and Xba I / 10 patterns derived from family B4 with DMD, and Xba I/4–5a pattern derived from family C1 with DMD. Particularly, both RFLPs and dosage intensity were detected in the pattern of Bgl II/5b-7 from family B4.
Three Bgl II patterns showing RFLPs and three Xba I patterns showing RFLPs in dystrophin cDNA hybridization with human genomic DNA from one BMD family(A1) and two DMD families (B4 and C1). Their pedigrees were shown in Fig 4, with arrows pointing to allele fragments. In the photograph from Bgl II /5b-7 in family B4, since III2 and III3 show deletion (del) with 3.5 kb, 3.3 kb and 2.8 kb fragments in hybridization pattern, the dosage intensity of the 3 bands was analysed in other 5 members. Referencing the normal pattern from normal adult male I1, used as a control, in which the intensity with 3.3 kb band is stronger than 3.4 kb band, the patterns from I2 and II2 were identified as complete dosage, while the patterns from II1 and III1 were identified as half dosage as 3.3 kb band intensity is less than 3.4 kb. “T” means the complete dosage, “H” means the half dosage.
Discussion
1. Number of Bgl II fragments
The normal Bgl II restriction fragment patterns were establised by using DNA samples taken from 20 normal males and 30 females of different ages . Except four new fragments, the Bgl II restriction patterns were similar to that of Caucacian19. Comparing the result of present work with the data reported previously19, we speculate that the four new fragments found in present study are unrelated with ethinic origin. The reasons are as follows: (1) when longer cDNA probes (>1.5 kb) were taken, the hybridization signal produced by shorter exons (<70 bp) can not usually be shown in patterns; when the shorter subprobe 1a (0.4 kb) was used, a new 6.2 kb fragment, which could not be shown in pattern of Bgl II/1–2a19, can be detected; (2) since the difference of size between two fragments is too small to be recognized in routine electrophoresis condition, a new 9.3 kb fragment in Bgl II/cDNA 9 pattern was identified after lengthening the electrophoresis of DNA samples; (3) since two or more than two fragments with similar molecular weights were shown as one band in some patterns, they were considered as one fragment in counting fragment number; the new 4.0 kb fragment was recognised by comparing hybridization patterns from 4 subprobes (11a, 11b, 11c–12a and 14) derived from cDNA 11–14; the new 3.7 kb fragment was identified by comparing 1a, 2a and 1–2a patterns too. Hence, in order to establish a partial map of the gene, it is necessary to prepare short subprobes by a strategy of overlap-cut probe and to lengthen the electrophoresis for DNA samples.
2. Rare polymorphic site
2 rare polymorphic sites in 2 RFLPs from the patterns of Bgl II/2b-3 and Bgl II /4–5a (Fig 1) were detected. The allelic fragments of two RFLPs reported by Darras and Francke in 198819 are as follows: (1) A1=23 kb, A3=8.5 kb in Bgl II/2b-3 pattern; (2) A1= 38.5 kb, A2=30+8.5 kb in Bgl II/4–5a pattern. However, an another 15 kb allelic fragment in (1) and a new 15 kb+8.5 kb allelic fragment in (2) described above were found in present study. They are two new rare polymorphsims. As shown in Fig 2, we regard the rare 15 kb fragment in Bgl II/2b-3 pattern to becaused by the disappearance of G2 site in Fig 2a and the rare 15 kb fragment in Bgl II/4–5a to be formed by a novel G3 site in Fig 2b.
3. Allelic frequency of Bgl II RFLPs and carrier testing
Comparing with the data from Caucacian19, there is no significant difference (Tab 2) in the frequency of four RFLPs in Chinese, although a RFLP from Bgl II/11–14 was not yet found in Chinese. The three RFLPs with high PIC value, can effectively be used in genetic counselling and prenatal diagnosis.
Since three Bgl II RFLPs can only detect about 1,000 kb in the central region of dystrophin gene11, 23, four Xba I RFLPs shown by cDNA 1–2a, 2b-3, 4–5a and 10, were used to extend the detected region to cover almost entire dystrophin gene including two hot-spot mutation regions. Three of four Xba I RFLPs were first revealed by our previous work20. Successful identification of carrier state for 29 females in 8 DMD/ BMD families was done by dosage intensity analysis and genetic linkage analysis with the 8 RFLPs distributed in entire dystrophin gene. In A2, B4, C1 and C2 families, the deletions were found in all 4 probands. 15 females from these families were accurately diagnosed with dosage intensity analysis. Five of them were defined as carrier of DMD mutation, 10 of them were normal. These results were consistent with those from linkage analysis. Although there is no deletion found in families A1 and B3, most of the individuals were still accurately diagnosed because most of these RFLPs showed allelic segragation. So 6 females were diagnosed as carrier of DMD, and 2 females as normal. But in families B1 and B2, the diagnosis was speculative because the allele-segragation of the RFLPs was limited.
In two females (II-1 in B2 family and II-1 in B3 family) of 40 offspring individuals, recombination occurred between Bgl II RFLP detected by cDNA 5b-7 and Xba I RFLP detected by cDNA 10. The recombination rate is 4 %. Fujishita et al (1991) 24 reported that the recombination between pERT 87–8 locus (against cDNA 2b-3) and 3′ end portion of the gene was found in 4 of 36 members in Japanese. and with a fraction of 11%. Abbs et al (1990)25 reported a fraction of 9.2 % in the two intragenic loci. This often makes it difficult to detect carriers accurately. Thus, more studies are needed to find novel RFLPs using dystrophin cDNA. Eight dinucleotide repeat (CA)n polymorphisms (three polymorphisms from 5′ end 26, 27, one from 3′ end28, and four from intron 44, 45, 49 and 50 of dystrophin gene29) were also used to detect the DMD/BMD carrier in our laboratory (to be published elsewhere), in which a part of the recombination within the gene can not be revealed for the dinucleotide repeat polymorphisms distributed with few and far between in the very large gene so that the accuracy of carrier diagnosis would be certainly affected. Moreover, 90 per cent of the deletion mutations can be found by multiplex DNA amplification with nine pairs of primers30 in 45 Chinese patients with DMD/BMD (to be published elsewhere), but carrier detection based on the product dosage of PCR amplification could not be carried out successfully. On the other hand, the presence of germinal mosaicism31 is also another problem for carrier detection. We think that rapid, economic, convenient, and accurate diagnosis for DMD/BMD carrier will depend on the combined application of several methods mentioned above accoding to the mutation type of proband with DMD or BMD. Therefore, gene dosage analysis with Southern hybridization is still important.
Recently, we have revealed 12 novel RFLPs (including 4 Taq I RFLPs, 3 Pvu II RFLPs, 2 EcoR V RFLPs and 3 Xba I RFLPs) by dystrophin cDNA, which will be published elsewhere20. Together with 26 RFLPs, which were also detected by dystrophin cDNA reported previously in Caucasian12, 14, 19, 32 and Japanese33, all 38 RFLPs distributed in entire gene will make the carrier testing based on dosage analysis and genotype analysis by dystrophin cDNA more sensitive and accurate.
References
Emery AEH . Duchenne muscular dystrophy. Oxford: Oxford University Press, 1986:151–156.
Francke U, Ochs HD, de Martinville B, et al. Minor Xp21 chromosome deletion in male associated with expression of Duchenne muscular dystrophy, chronic granulomatous disease, retinitis pigmentosa, and McLeod syndrome. Am J Hum Genet 1985; 37: 250–67.
Boyd Y, Buckle VJ . Cytogenetic heterogeneity of translocations associated with Duchenne muscular dystrophy. Clin Genet 1986; 29:108–15.
Davies KE, Pearson PL, Harper PS, Murray JM, O'Brien T, Sarfarazi M, et al. Linkage analysis of two cloned DNA sequences flanking the Duchenne muscular dystrophy locus on the short arm of the human X chromosome. Nucleic Acids Res 1983; 11:2303–12.
Kingston HM, Sarfarazi M, Thomas NST, Harper PS . Localization of the Becker muscular dystrophy gene on the short arm of the X chromosome by linkage to cloned DNA sequences. Hum Genet 1984; 67:6–17.
Kunkel LM, Monaco AP, Middlesworth W, Ochs HD, Latt SA . Specific cloning of DNA fragments absent from the DNA of a male patient with an X chromosome deletion. Proc Natl Acad Sci USA 1985; 82:4778–82.
Monaco AP, Bertelson C J, Middlesworth W, Colletti C, Aldridge J, Fischbeck KH, et al. Detection of deletions spanning the Duchenne muscular dystrophy locus using a tightly linked DNA segment. Nature 1985; 316:842–5.
Kunkel LM and 76 co-authors. Analysis of deletions in DNA from patients with Becker and Duchenne muscular dystrophy. Nature 1986; 322:73–7.
Ray PN, Belfall B, Duff C, Logan C, Kean V, Thompson MW, et al. Cloning of the breakpoint of an Xp21 translocation associated with Duchenne muscular dystrophy. Nature 1985; 318:672–5.
Monaco AP, Verteleson CJ, Colletti-Feener C, Kunkel LM . Localization and cloning of Xp21 deletion breakpoints involved in muscular dystrophy, Hum Genet 1987; 75:221–7.
den Dunnen JT, Barkker E, Klein Breteler EG, Pearson PL, van Ommen GJB . Direct detection of more than 50% of the Duchenne muscular dystrophy mutations by field inversion gels. Nature 1987; 329:640–2.
Koenig M, Hoffman EP, Bertelson C J, Monaco AP, Feener C, Kunkel LM . Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell 1987; 50:509–17.
Koenig M, Beggs AH, Moyer M, Scherpf S, Heindrich K, Bettecken T, et al. The molecular basis for Duchenne versus Becker muscular dystrophy correlation of severity with type of deletion. Am J Hum Genet 1989; 48:498–506.
Forrest S, Smith YJ, Cross GS, et al. Effective strategy for prenatal prediction of Duchenne and Becker muscular dystrophy. Lancet 1987; 2:1294–7.
Darras BT, Blattner P, Harper JF, Spiro A J, Alter S, Francke U . Intragenic deletions in 21 Duchenne muscular dystrophy (DMD)/ Becker muscular dystrophy (BMD) families studied with the dystrophin cDNA: location of breakpoints on Hind III and Bgl II exon-containing fragment maps, meiotic and mitotic origin of the mutations. Am J Hum Genet 1988; 43:620–9.
Yu L, Wang N, Hua XY, Cai T, Zhang XX, Yang YM, et al. Topography of molecular rearrangement of dystrophin gene in Chinese: deletion and duplication detection in 123 cases with DMD or BMD. (In preparation)
Roberts RG, Cole CG, Hart KA, Bobrow M, Bently DR . Rapid carrier and prenatal diagnosis of Duchenne and Becker muscular dystrophy. Nucleic Acids Res 1989; 17:811.
Mao YP, Cremer M . Deletion of Duchenne muscular dystrophy carriers by dosage analysis using the DMD cDNA clone 8. Human Genet 1989; 81:193–5.
Darras BT, Francke U . Normal human genomic restriction fragment patterns and polymorphisms revealed by hybridization with the entire dystrophin cDNA. Am J Hum Genet 1988; 43:612–9.
Yu L, Wang N, Zhao SY . Twelve novel polymorphisms for dystrophin gene in Chinese. (In preparation)
Kunkel LM, Smith KD, Boyer SH . Analysis of human Y-chromosome specific reiterated DNA in chromosome variants. Proc Natl Acad Sci USA 1977; 174:1245–9.
Yu L, Wang MQ, Wang QB, Wang WY, Yang YM, Zhu JD, Zhao SY . Comparing the frequencies of restriction fragment length polymorphisms for dystrophin gene in Chinese with those from Japanes and Caucasian populations. Cell Research 1993; 3:39–47.
Burmeister M, Monaco AP, Gillard EF, van Ommen GJB, Affara NA, Ferguson-smith MA, et al. A 10-megabase physical map of human Xp21, including the Duchenne muscular dystrophy gene. Genomics 1988; 2:189–202.
Fujishita S, Shibuya N, Niikawa N, Nagataki S . Gene-deletion and carrier detection, and prenatal diagnosis of Duchenne muscular dystrophy by analysis of the dystrophies gene amplified by PCR. Jpn J Hum Genet 1991; 36:317–24.
Abbs S, Robert RG, Mothew GG, Bently DR and Bobrow M . Accurate assessment of intragenic recombination frequency within the Duchenne muscular dystrophy gene. Genomics 1990; 7:602–6.
Oudet C, Heilig R, Hanauer A and Mandel JL . Nonradioactive assay for new microsatellite polymorphisms at the 5′ end of the dystrophin gene,and estimation of introgenic recombination. Am J Hum Genet 1991; 49:311–9.
Feener CA, Boyce FM and Kunkel LM . Rapid detection of CA polymorphisms in cloned DNA: application to the 5′ region of the dystrophin gene. Am J Hum Genet 1991; 48:621–7.
Clemens PR, Fenwick RG, Chamberlain JS, Gibbs RA, Andrade M, Chakraborty R and Caskey CT . Carrier detection and prenatal diagnosis in Duchenne and Becker muscular dystrophy families, using dinucleotide repeat polymorphisms. Am J Hum Genet 1991; 49:951–60.
Oudet C, Heilig R and Mandel JL . An informative polymorphism detectable by polymerase chain reaction at the 3' end of the dystrophin gene. Hum Genet 1990; 84:283–5.
Chamberlain JS, Gibbs RA, Ranier JE, Nguyen PN, Caskey CT . Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res 1988; 16:11141–56.
Bakker E, van Broeckhoven C, Bontern E J, van de Vooren M J, Veenema H, Van Hul W, et al. Germline mosaicism and Duchenne muscular dystrophy mutation. Nature 1987; 329:554–6.
Gallati SL, Schneider V, Mullis P, and Moser H . RFLPs for Duchenne muscular dystrophy cDNA clone 9 and 10. Am J Hum Genet 1990; 46:1090–4.
Deng HX and Niikawa N . Pvu II RFLPs in the DMD gene detected by a subclone (cDMD 1a) of the cDNA. Nucleic Acids Res 1990; 16:9072.
Acknowledgements
The authors thank Prof. Liu TT and Dr. Cai T for scientific discussion. This work is supported by National Natural Science Foundation of China, the Foundation of Shanghai Science and Technology Commission and Shanghai Joint Laboratory of Life Sciences, Academia Sinica.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Yu, L., Wang, N., Deng, Y. et al. Structural analysis of DMD gene and its clinical application in Chinese.I.Bgl II exon-containing fragment, RFLP and carrier detection. Cell Res 4, 201–215 (1994). https://doi.org/10.1038/cr.1994.21
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/cr.1994.21