Abstract
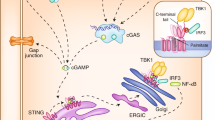
Although it has long been demonstrated that cytosolic DNA is a potent immune stimulant, it is only in recent years that the molecular mechanisms of DNA-stimulated innate immune responses have emerged. Studies have established critical roles for the DNA sensor cyclic GMP–AMP synthase (cGAS) and the adapter protein MITA/STING in the innate immune response to cytosolic DNA or DNA viruses. Although the regulation of cGAS–MITA/STING-mediated signaling remains to be fully investigated, understanding the processes involved may help to explain the mechanisms of innate immune signaling events and perhaps autoinflammatory diseases and to provide potential therapeutic targets for drug intervention. In this review, we summarize recent progress on the regulation of the cGAS–MITA/STING-mediated innate immune response to DNA viruses at the organelle-trafficking, post-translational and transcriptional levels.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell 2010; 140: 805–820.
Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol 2004; 4: 499–511.
Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity 2010; 32: 305–315.
Oldenburg M, Kruger A, Ferstl R, Kaufmann A, Nees G, Sigmund A et al. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science 2012; 337: 1111–1115.
Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 2007; 448: 501–505.
Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol 2011; 12: 959–965.
Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 2010; 11: 997–1004.
Xia P, Wang S, Ye B, Du Y, Huang G, Zhu P et al. Sox2 functions as a sequence-specific DNA sensor in neutrophils to initiate innate immunity against microbial infection. Nat Immunol 2015; 16: 366–375.
Li Y, Chen R, Zhou Q, Xu Z, Li C, Wang S et al. LSm14A is a processing body-associated sensor of viral nucleic acids that initiates cellular antiviral response in the early phase of viral infection. Proc Natl Acad Sci USA 2012; 109: 11770–11775.
Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 2009; 458: 509–513.
Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 2009; 458: 514–518.
Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol 2009; 10: 266–272.
Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science 2009; 323: 1057–1060.
Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 2008; 455: 674–678.
Sun W, Li Y, Chen L, Chen H, You F, Zhou X et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proce Natl Acad Sci USA 2009; 106: 8653–8658.
Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. MPY, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cel Biol 2008; 28: 5014–5026.
Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 2008; 29: 538–550.
Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 2011; 478: 515–518.
Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 2010; 328: 1703–1705.
Wu J, Sun L, Chen X, Du F, Shi H, Chen C et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 2013; 339: 826–830.
Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013; 339: 786–791.
Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 2013; 341: 1390–1394.
Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 2009; 461: 788–792.
Baguley BC, Ching LM. DMXAA: an antivascular agent with multiple host responses. Int J Radiat Oncol Biol Phys 2002; 54: 1503–1511.
Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov AA et al. Structure-function analysis of STING activation by c[G(2',5')pA(3',5')p] and targeting by antiviral DMXAA. Cell 2013; 154: 748–762.
Prantner D, Perkins DJ, Lai W, Williams MS, Sharma S, Fitzgerald KA et al. 5,6-Dimethylxanthenone-4-acetic acid (DMXAA) activates stimulator of interferon gene (STING)-dependent innate immune pathways and is regulated by mitochondrial membrane potential. J Biol Chem 2012; 287: 39776–39788.
Holm CK, Jensen SB, Jakobsen MR, Cheshenko N, Horan KA, Moeller HB et al. Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat Immunol 2012; 13: 737–743.
Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M et al. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep 2013; 3: 1355–1361.
Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I et al. cGAS produces a 2'-5'-linked cyclic dinucleotide second messenger that activates STING. Nature 2013; 498: 380–384.
Schaap P. Cyclic di-nucleotide signaling enters the eukaryote domain. IUBMB life 2013; 65: 897–903.
Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol 2011; 30: 16–34.
Franchi L, Munoz-Planillo R, Nunez G. Sensing and reacting to microbes through the inflammasomes. Nat Immunol 2012; 13: 325–332.
Ahn J, Gutman D, Saijo S, Barber GN. STING manifests self DNA-dependent inflammatory disease. Proc Natl Acad Sci USA 2012; 109: 19386–19391.
Corrales L, Woo SR, Williams JB, McWhirter SM, Dubensky Jr TW, Gajewski TF. Antagonism of the STING Pathway via Activation of the AIM2 Inflammasome by Intracellular DNA. J Immunol 2016; 196: 3191–3198.
Chen H, Sun H, You F, Sun W, Zhou X, Chen L et al. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell 2011; 147: 436–446.
Dong G, You M, Ding L, Fan H, Liu F, Ren D et al. STING Negatively Regulates Double-Stranded DNA-Activated JAK1-STAT1 Signaling via SHP-1/2 in B Cells. Mol Cell 2015; 38: 441–451.
Zhong B, Zhang L, Lei C, Li Y, Mao AP, Yang Y et al. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 2009; 30: 397–407.
Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D et al. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog 2012; 8: e1002934.
You F, Wang P, Yang L, Yang G, Zhao YO, Qian F et al. ELF4 is critical for induction of type I interferon and the host antiviral response. Nat Immunol 2013; 14: 1237–1246.
Nazmi A, Mukhopadhyay R, Dutta K, Basu A. STING mediates neuronal innate immune response following Japanese encephalitis virus infection. Sci Rep 2012; 2: 347.
Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 2014; 505: 691–695.
Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 2011; 472: 481–485.
Lim JK, Lisco A, McDermott DH, Huynh L, Ward JM, Johnson B et al. Genetic variation in OAS1 is a risk factor for initial infection with West Nile virus in man. PLoS Pathog 2009; 5: e1000321.
Martinez Valle F, Balada E, Ordi-Ros J, Vilardell-Tarres M. DNase 1 and systemic lupus erythematosus. Autoimmun Rev 2008; 7: 359–363.
Kawane K, Fukuyama H, Kondoh G, Takeda J, Ohsawa Y, Uchiyama Y et al. Requirement of DNase II for definitive erythropoiesis in the mouse fetal liver. Science 2001; 292: 1546–1549.
Kawane K, Fukuyama H, Yoshida H, Nagase H, Ohsawa Y, Uchiyama Y et al. Impaired thymic development in mouse embryos deficient in apoptotic DNA degradation. Nat Immunol 2003; 4: 138–144.
Morita M, Stamp G, Robins P, Dulic A, Rosewell I, Hrivnak G et al. Gene-targeted mice lacking the Trex1 (DNase III) 3'—>5' DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol 2004; 24: 6719–6727.
Kawane K, Ohtani M, Miwa K, Kizawa T, Kanbara Y, Yoshioka Y et al. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature 2006; 443: 998–1002.
Gao D, Li T, Li XD, Chen X, Li QZ, Wight-Carter M et al. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc Natl Acad Sci USA 2015; 112: E5699–E5705.
Rice GI, Rodero MP, Crow YJ. Human disease phenotypes associated with mutations in TREX1. J Clin Immunol 2015; 35: 235–243.
Gall A, Treuting P, Elkon KB, Loo YM, Gale Jr M, Barber GN et al. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity 2012; 36: 120–131.
Ablasser A, Hemmerling I, Schmid-Burgk JL, Behrendt R, Roers A, Hornung V. TREX1 deficiency triggers cell-autonomous immunity in a cGAS-dependent manner. J Immunol 2014; 192: 5993–5997.
Gray EE, Treuting PM, Woodward JJ, Stetson DB. Cutting edge: cGAS is required for lethal autoimmune disease in the Trex1-deficient mouse model of Aicardi-Goutieres syndrome. J Immunol 2015; 195: 1939–1943.
Pokatayev V, Hasin N, Chon H, Cerritelli SM, Sakhuja K, Ward JM et al. RNase H2 catalytic core Aicardi-Goutieres syndrome-related mutant invokes cGAS-STING innate immune-sensing pathway in mice. J Exp Med 2016; 213: 329–336.
Mackenzie KJ, Carroll P, Lettice L, Tarnauskaite Z, Reddy K, Dix F et al. Ribonuclease H2 mutations induce a cGAS/STING-dependent innate immune response. EMBO J 2016; 35: 831–844.
Liu Y, Jesus AA, Marrero B, Yang D, Ramsey SE, Montealegre Sanchez GA et al. Activated STING in a vascular and pulmonary syndrome. New Engl J Med 2014; 371: 507–518.
Jeremiah N, Neven B, Gentili M, Callebaut I, Maschalidi S, Stolzenberg MC et al. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J Clin Invest 2014; 124: 5516–5520.
Chien Y, Kim S, Bumeister R, Loo YM, Kwon SW, Johnson CL et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell 2006; 127: 157–170.
Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 2013; 155: 688–698.
Dobbs N, Burnaevskiy N, Chen D, Gonugunta VK, Alto NM, Yan N. STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host Microbe 2015; 18: 157–168.
Chen W, Li S, Yu H, Liu X, Huang L, Wang Q et al. ER Adaptor SCAP Translocates and Recruits IRF3 to Perinuclear Microsome Induced by Cytosolic Microbial DNAs. PLoS Pathog 2016; 12: e1005462.
Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci USA 2009; 106: 20842–20846.
Liang Q, Seo GJ, Choi YJ, Kwak MJ, Ge J, Rodgers MA et al. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe 2014; 15: 228–238.
Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity 2012; 37: 223–234.
Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol 2009; 10: 1215–1221.
Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 2012; 150: 803–815.
Rasmussen SB, Horan KA, Holm CK, Stranks AJ, Mettenleiter TC, Simon AK et al. Activation of autophagy by alpha-herpesviruses in myeloid cells is mediated by cytoplasmic viral DNA through a mechanism dependent on stimulator of IFN genes. J Immunol 2011; 187: 5268–5276.
McIlwain DR, Lang PA, Maretzky T, Hamada K, Ohishi K, Maney SK et al. iRhom2 regulation of TACE controls TNF-mediated protection against Listeria and responses to LPS. Science 2012; 335: 229–232.
Siggs OM, Xiao N, Wang Y, Shi H, Tomisato W, Li X et al. iRhom2 is required for the secretion of mouse TNFalpha. Blood 2012; 119: 5769–5771.
Luo WW, Li S, Li C, Lian H, Zhong B, Shu HB. iRhom2 is essential for innate immunity to DNA viruses by mediating trafficking and stability of the adaptor STING. Nat Immunol 2016; 17: 1057–1066.
Issuree PD, Maretzky T, McIlwain DR, Monette S, Qing X, Lang PA et al. iRHOM2 is a critical pathogenic mediator of inflammatory arthritis. J Clin Invest 2013; 123: 928–932.
Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y et al. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity 2010; 33: 765–776.
Zhang J, Hu MM, Wang YY, Shu HB. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem 2012; 287: 28646–28655.
Wang Q, Liu X, Cui Y, Tang Y, Chen W, Li S et al. The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity 2014; 41: 919–933.
Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity 2012; 36: 1073–1086.
Zhong B, Zhang Y, Tan B, Liu TT, Wang YY, Shu HB. The E3 ubiquitin ligase RNF5 targets virus-induced signaling adaptor for ubiquitination and degradation. J Immunol 2010; 184: 6249–6255.
Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell 2005; 19: 727–740.
Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 2005; 122: 669–682.
Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 2005; 6: 981–988.
Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 2005; 437: 1167–1172.
Wang Y, Lian Q, Yang B, Yan S, Zhou H, He L et al. TRIM30alpha is a negative-feedback regulator of the intracellular DNA and DNA virus-triggered response by targeting STING. PLoS Pathog 2015; 11: e1005012.
Qin Y, Zhou MT, Hu MM, Hu YH, Zhang J, Guo L et al. RNF26 temporally regulates virus-triggered type I interferon induction by two distinct mechanisms. PLoS Pathog 2014; 10: e1004358.
Hu MM, Yang Q, Xie XQ, Liao CY, Lin H, Liu TT et al Sumoylation Promotes the Stability of the DNA Sensor cGAS and the Adaptor STING to Regulate the Kinetics of Response to DNA Virus. Immunity 2016; 45: 555–569.
Ran Y, Liu TT, Zhou Q, Li S, Mao AP, Li Y et al. SENP2 negatively regulates cellular antiviral response by deSUMOylating IRF3 and conditioning it for ubiquitination and degradation. J Mol Cell Biol 2011; 3: 283–292.
Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal 2012; 5: ra20.
Li Z, Liu G, Sun L, Teng Y, Guo X, Jia J et al. PPM1A regulates antiviral signaling by antagonizing TBK1-mediated STING phosphorylation and aggregation. PLoS Pathog 2015; 11: e1004783.
Lei CQ, Zhong B, Zhang Y, Zhang J, Wang S, Shu HB. Glycogen synthase kinase 3beta regulates IRF3 transcription factor-mediated antiviral response via activation of the kinase TBK1. Immunity 2010; 33: 878–889.
Liu S, Cai X, Wu J, Cong Q, Chen X, Li T et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 2015; 347: aaa2630.
Seo GJ, Yang A, Tan B, Kim S, Liang Q, Choi Y et al. Akt kinase-mediated checkpoint of cGAS DNA sensing pathway. Cell Rep 2015; 13: 440–449.
Janke C, Rogowski K, Wloga D, Regnard C, Kajava AV, Strub JM et al. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science 2005; 308: 1758–1762.
Rogowski K, van Dijk J, Magiera MM, Bosc C, Deloulme JC, Bosson A et al. A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell 2010; 143: 564–578.
Garnham CP, Vemu A, Wilson-Kubalek EM, Yu I, Szyk A, Lander GC et al. Multivalent Microtubule Recognition by Tubulin Tyrosine Ligase-like Family Glutamylases. Cell 2015; 161: 1112–1123.
Janke C, Rogowski K, van Dijk J. Polyglutamylation: a fine-regulator of protein function? 'Protein Modifications: beyond the usual suspects' review series. EMBO Rep 2008; 9: 636–641.
Xia P, Ye B, Wang S, Zhu X, Du Y, Xiong Z et al. Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nat Immunol 2016; 17: 369–378.
Rodriguez de la Vega, Otazo M, Lorenzo J, Tort O, Aviles FX, Bautista JM. Functional segregation and emerging role of cilia-related cytosolic carboxypeptidases (CCPs). FASEB J 2013; 27: 424–431.
Berezniuk I, Sironi J, Callaway MB, Castro LM, Hirata IY, Ferro ES et al. CCP1/Nna1 functions in protein turnover in mouse brain: Implications for cell death in Purkinje cell degeneration mice. FASEB J 2010; 24: 1813–1823.
Ma F, Li B, Liu SY, Iyer SS, Yu Y, Wu A et al. Positive feedback regulation of type I IFN production by the IFN-inducible DNA sensor cGAS. J Immunol 2015; 194: 1545–1554.
Panchanathan R, Liu H, Xin D, Choubey D. Identification of a negative feedback loop between cyclic di-GMP-induced levels of IFI16 and p202 cytosolic DNA sensors and STING. Innate Immu 2014; 20: 751–759.
Liu TT, Yang Q, Li M, Zhong B, Ran Y, Liu LL et al. LSm14A plays a critical role in antiviral immune responses by regulating MITA level in a cell-specific manner. J Immunol 2016; 196: 5101–5111.
Yarbrough ML, Zhang K, Sakthivel R, Forst CV, Posner BA, Barber GN et al. Primate-specific miR-576-3p sets host defense signalling threshold. Nat Commun 2014; 5: 4963.
Zhang L, Mo J, Swanson KV, Wen H, Petrucelli A, Gregory SM et al. NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity 2014; 40: 329–341.
Guo H, Konig R, Deng M, Riess M, Mo J, Zhang L et al. NLRX1 Sequesters STING to Negatively Regulate the Interferon Response, Thereby Facilitating the Replication of HIV-1 and DNA Viruses. Cell Host Microbe 2016; 19: 515–528.
Barouch DH, Ghneim K, Bosche WJ, Li Y, Berkemeier B, Hull M et al. Rapid inflammasome activation following mucosal SIV infection of Rhesus monkeys. Cell 2016; 165: 656–667.
Zhou Q, Lin H, Wang S, Wang S, Ran Y, Liu Y et al. The ER-associated protein ZDHHC1 is a positive regulator of DNA virus-triggered, MITA/STING-dependent innate immune signaling. Cell Host Microbe 2014; 16: 450–461.
Ma Z, Damania B. The cGAS-STING defense pathway and its counteraction by viruses. Cell Host Microbe 2016; 19: 150–158.
Christensen MH, Paludan SR. Viral evasion of DNA-stimulated innate immune responses. Cell Mol Immunol; 2016 e-pub ahead of print 14 March 2016, doi:10.1038/cmi.2016.06.
Acknowledgements
The work in the authors’ laboratory is supported by grants from the Ministry of Science and Technology of China (2014CB910103, 2012CB910201 and 2016YFA0502100) and the National Natural Science Foundation of China (3163000013, 31521091 and 91429304).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Luo, WW., Shu, HB. Delicate regulation of the cGAS–MITA-mediated innate immune response. Cell Mol Immunol 15, 666–675 (2018). https://doi.org/10.1038/cmi.2016.51
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cmi.2016.51
Keywords
This article is cited by
-
Mitochondrial DNA-triggered innate immune response: mechanisms and diseases
Cellular & Molecular Immunology (2023)
-
Clinical significance of STING expression and methylation in lung adenocarcinoma based on bioinformatics analysis
Scientific Reports (2022)
-
The RNA-binding protein LUC7L2 mediates MITA/STING intron retention to negatively regulate innate antiviral response
Cell Discovery (2021)
-
SNX8 modulates the innate immune response to RNA viruses by regulating the aggregation of VISA
Cellular & Molecular Immunology (2020)
-
DNA-PK deficiency potentiates cGAS-mediated antiviral innate immunity
Nature Communications (2020)