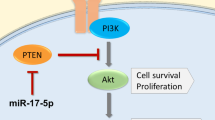
Abstract
We examined the microRNA (miRNA) expression profile of 40 prostatectomy specimens from stage T2a/b, early relapse and non-relapse cancer patients, to better understand the relationship between miRNA dysregulation and prostate oncogenesis. Paired analysis was carried out with microdissected, malignant and non-involved areas of each specimen, using high-throughput liquid-phase hybridization (mirMASA) reactions and 114 miRNA probes. Five miRNAs (miR-23b, -100, -145, -221 and -222) were significantly downregulated in malignant tissues, according to significance analysis of microarrays and paired t-test with Bonferroni correction. Lowered expression of miR-23b, -145, -221 and -222 in malignant tissues was validated by quantitative reverse transcription (qRT)-PCR analyses. Ectopic expression of these miRNAs significantly reduced LNCaP cancer cell growth, suggesting growth modulatory roles for these miRNAs. Patient subset analysis showed that those with post-surgery elevation of prostate-specific antigen (chemical relapse) displayed a distinct expression profile of 16 miRNAs, as compared with patients with non-relapse disease. A trend of increased expression (>40%) of miR-135b and miR-194 was observed by qRT-PCR confirmatory analysis of 11 patients from each clinical subset. These findings indicate that an altered miRNA expression signature accompanied the prostate oncogenic process. Additional, aberrant miRNA expression features may reflect a tendency for early disease relapse. Growth inhibition through the reconstitution of miRNAs is potentially applicable for experimental therapy of prostate cancer, pending molecular validation of targeted genes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lee RC, Feinbaum RL, Ambros V . The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75: 843–854.
Lau NC, Lim LP, Weinstein EG, Bartel DP . An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001; 294: 858–862.
Ambros V . MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 2003; 113: 673–676.
Harfe BD . MicroRNAs in vertebrate development. Curr Opin Genet Dev 2005; 15: 410–415.
Xi Y, Shalgi R, Fodstad O, Pilpel Y, Ju J . Differentially regulated micro-RNAs and actively translated messenger RNA transcripts by tumor suppressor p53 in colon cancer. Clin Cancer Res 2006; 12 (7 Part 1): 2014–2024.
Mathupala SP, Guthikonda M, Sloan AE . RNAi based approaches to the treatment of malignant glioma. Technol Cancer Res Treat 2006; 5: 261–269.
Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 2004; 101: 2999–3004.
Gong H, Liu CM, Liu DP, Liang CC . The role of small RNAs in human diseases: potential troublemaker and therapeutic tools. Med Res Rev 2005; 25: 361–381.
Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A et al. RAS is regulated by the let-7 microRNA family. Cell 2005; 120: 635–647.
Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA 2005; 102: 13944–13949.
Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K et al. MicroRNAs modulate the angiogenic properties of HUVECs. Blood 2006; 108: 3068–3071.
Tong AW, Nemunaitis J . Modulation of miRNA activity in human cancer: a new paradigm for cancer gene therapy? Cancer Gene Ther 2008; 15: 341–355.
O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT . c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005; 435: 839–843.
American Cancer Society. Cancer Facts and Figures 2008. Atlanta: American Cancer Society 2008.
Walczak JR, Carducci MA . Prostate cancer: a practical approach to current management of recurrent disease. Mayo Clin Proc 2007; 82: 243–249.
Bradford TJ, Tomlins SA, Wang X, Chinnaiyan AM . Molecular markers of prostate cancer. Urol Oncol 2006; 24: 538–551.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435: 834–838.
Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 2006; 103: 2257–2261.
Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005; 65: 7065–7070.
Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene 2006; 25: 2537–2545.
Yang L, Tran DK, Wang X . BADGE, beads array for the detection of gene expression, a high-throughput diagnostic bioassay. Genome Res 2001; 11: 1888–1898.
Petersen M, Wengel J . LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol 2003; 21: 74–81.
Barad O, Meiri E, Avniel A, Aharonov R, Barzilai A, Bentwich I et al. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res 2004; 14: 2486–2494.
Tusher VG, Tibshirani R, Chu G . Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001; 98: 5116–5121.
Zhang YA, Nemunaitis J, Scanlon KJ, Tong AW . Anti-tumorigenic effect of a K-ras ribozyme against human lung cancer cell line heterotransplants in nude mice. Gene Therapy 2000; 7: 2041–2050.
Jay C, Nemunaitis J, Chen P, Fulgham P, Tong AW . miRNA profiling for diagnosis and prognosis of human cancer. DNA Cell Biol 2007; 26: 293–300.
Nelson PT, Baldwin DA, Scearce LM, Oberholtzer JC, Tobias JW, Mourelatos Z . Microarray-based, high-throughput gene expression profiling of microRNAs. Nat Methods 2004; 1: 155–161.
van Rooij E, Olson EN . MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest 2007; 117: 2369–2376.
Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005; 433: 769–773.
Bandres E, Cubedo E, Agirre X, Malumbres R, Zarate R, Ramirez N et al. Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer 2006; 5: 29.
Rana TM . Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol 2007; 8: 23–36.
Coller HA, Forman JJ, Legesse-Miller A . ‘Myc’ed messages’: myc induces transcription of E2F1 while inhibiting its translation via a microRNA polycistron. PLoS Genet 2007; 3: e146.
White RJ . RNA polymerase III transcription and cancer. Oncogene 2004; 23: 3208–3216.
Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer 2006; 5: 24.
Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T . MicroRNA expression profiling in prostate cancer. Cancer Res 2007; 67: 6130–6135.
Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafre SA et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem 2007; 282: 23716–23724.
Ozen M, Creighton CJ, Ozdemir M, Ittmann M . Widespread deregulation of microRNA expression in human prostate cancer. Oncogene 2008; 27: 1788–1793.
Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Fabio P et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res 2008; 68: 6162–6170.
Diederichs S, Haber DA . Sequence variations of microRNAs in human cancer: alterations in predicted secondary structure do not affect processing. Cancer Res 2006; 66: 6097–6104.
Michael MZ, O’Connor SM, van Holst Pellekaan NG, Young GP, James RJ . Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res 2003; 1: 882–891.
Cummins JM, Velculescu VE . Implications of micro-RNA profiling for cancer diagnosis. Oncogene 2006; 25: 6220–6227.
le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J 2007; 26: 3699–3708.
Pallante P, Visone R, Ferracin M, Ferraro A, Berlingieri MT, Troncone G et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer 2006; 13: 497–508.
Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG . Regulation of miRNA expression during neural cell specification. Eur J Neurosci 2005; 21: 1469–1477.
Kawasaki H, Taira K . Functional analysis of microRNAs during the retinoic acid-induced neuronal differentiation of human NT2 cells. Nucleic Acids Res Suppl 2003; 3: 243–244.
Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F et al. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem 2007; 282: 2135–2143.
Lin SL, Chiang A, Chang D, Ying SY . Loss of mir-146a function in hormone-refractory prostate cancer. RNA 2008; 14: 417–424.
Weinstein IB . Cancer. Addiction to oncogenes—the Achilles heal of cancer. Science 2002; 297: 63–64.
Hino K, Tsuchiya K, Fukao T, Kiga K, Okamoto R, Kanai T et al. Inducible expression of microRNA-194 is regulated by HNF-1alpha during intestinal epithelial cell differentiation. RNA 2008; 14: 1433–1442.
Koturbash I, Boyko A, Rodriguez-Juarez R, McDonald RJ, Tryndyak VP, Kovalchuk I et al. Role of epigenetic effectors in maintenance of the long-term persistent bystander effect in spleen in vivo. Carcinogenesis 2007; 28: 1831–1838.
Acknowledgements
We thank Dr Qunying Yang at Genaco Biomedical, Huntsville, AL, for her technical assistance in mirMASA analyses; Ms Angela Clark at Presbysterian Hospital, Dallas, TX, for the generation and management of the patient database and Drs Damian Chaussabel and Casey Glaser at the Baylor Institute for Immunological Research, Dallas, TX, for their insights and suggestions on SAM. This study was supported in part by the Mary Crowley Research Fund.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Cancer Gene Therapy website (http://www.nature.com/cgt)
Rights and permissions
About this article
Cite this article
Tong, A., Fulgham, P., Jay, C. et al. MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther 16, 206–216 (2009). https://doi.org/10.1038/cgt.2008.77
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cgt.2008.77
Keywords
This article is cited by
-
Pro-tumorigenic role of lnc-ZNF30-3 as a sponge counteracting miR-145-5p in prostate cancer
Biology Direct (2023)
-
miR-32 promotes MYC-driven prostate cancer
Oncogenesis (2022)
-
MiRNAs in renal cell carcinoma
Clinical and Translational Oncology (2022)
-
Validation of the four-miRNA biomarker panel MiCaP for prediction of long-term prostate cancer outcome
Scientific Reports (2020)
-
Deregulated microRNAs in neurofibromatosis type 1 derived malignant peripheral nerve sheath tumors
Scientific Reports (2020)