Abstract
Jumonji C (JmjC) domain-containing protein 14 (JMJ14) is an H3K4-specific histone demethylase that has important roles in RNA-mediated gene silencing and flowering time regulation in Arabidopsis. However, how JMJ14 is recruited to its target genes remains unclear. Here, we show that the C-terminal FYRN (F/Y-rich N terminus) and FYRC (F/Y-rich C terminus) domains of JMJ14 are required for RNA silencing and flowering time regulation. Chromatin binding of JMJ14 is lost upon deletion of its FYRN and FYRC domains, and H3K4me3 is increased. FYRN and FYRC domains interact with a pair of NAC (NAM, ATAF, CUC) domain-containing transcription factors, NAC050 and NAC052. Genome-wide chromatin immunoprecipitation analysis revealed that JMJ14 and NAC050/052 share a set of common target genes with CTTGNNNNNCAAG consensus sequences. Mutations in either NAC052 or NAC050 impair RNA-mediated gene silencing. Together, our findings demonstrate an important role of FYRN and FYRC domains in targeting JMJ14 through direct interaction with NAC050/052 proteins, which reveals a novel mechanism of histone demethylase recruitment.
Similar content being viewed by others
Introduction
Histone lysine methylation has important roles in diverse biological functions including heterochromatin formation, transcriptional gene silencing, transcriptional activation, DNA repair, and DNA recombination [1–3]. Histone methylation can be removed by Jumonji C (JmjC) domain-containing proteins (JMJs) and LSD1 family proteins [4]. In Arabidopsis, some JmjC domain-containing proteins have been shown to remove methyl groups from different lysine residues of histone H3, thereby participating in controlling transcriptional repression and DNA methylation for genome stability, and in regulation of plant development [5–16]. JmjC domain-containing protein 14 (JMJ14) has been demonstrated to demethylate tri-, di-, and mono-methylated H3K4 [9–11]. JMJ14 contributes to flowering time regulation through repression of the florigen FLOWERING LOCUS T (FT), the floral integrators APETALA1 (AP1), SUPPRESSOR OF CO OVEREXPRESSION 1 (SOC1), and LFY [10]. Additional lines of evidence indicate that JMJ14 is also involved in transgene silencing that acts on cell-to-cell movement of an RNA silencing signal, reduction of transgene transcription, maintenance of endogenous transposon silencing, and DRM2-mediated CHH DNA methylation [12, 14, 16]. jmj14 mutants suppress post-transcriptional gene silencing at a variety of targets, including JAP3 loci carrying a pSUC2-driven PHYTOENE DESATURASE (PDS) inverted repeat transgene and a 35S-driven β-glucuronidase transgene (L1) undergoing sense transgene silencing (S-post-transcriptional gene silencing) [12, 16]. In jmj14 mutants, transcription levels of transgene loci were downregulated, whereas several known endogenous targets of JMJ14 are upregulated [16].
JMJ14 belongs to the KDM5/JARID1 subfamily of JmjC proteins, in which the plant and animal counterparts contain distinct domains: animal and plant proteins contain JmjN, JmjC, and C5HC2 zinc-finger domains, while the AT-rich interaction domain (ARID) and plant homeodomain (PHD) domains existing in members of yeast and animals are missing in most of the plant proteins [17]. There are several PHD domain subtypes in mammals and yeast, among which the first PHD domain of Jarid1C in human and the second PHD domain of Lid2 in yeast were reported to recognize methylated H3K9 [18, 19]. In Arabidopsis, all active KDM5 members, including JMJ14, JMJ15, JMJ16 and JMJ18–lack PHD domains, but contain an F/Y-rich N terminus (FYRN) and an F/Y-rich C terminus (FYRC) domains in the C terminus [2, 10, 17, 20]. FYRN and FYRC domains are rich in phenylalanine/tyrosine residues, which are found in many chromatin-related proteins, such as mixed lineage leukemia 1 and mixed lineage leukemia 2, two histone H3K4 methyltransferases in human [21, 22]/nuclear interactor of ARF and MDM2, a growth inhibitory protein involved in maintaining chromosomal stability [23], ATX1, an H3K4 methyltransferase involved in plant development and stress responses [24, 25], and AtMBD9, a methyl-CpG binding domain-containing protein involved in plant development [26].
In this study, we find that FYRN and FYRC domains are essential for JMJ14 function. These two domains have critical role in JMJ14 binding to its genomic targets through interacting with transcription factors NAC050 and NAC052. Thus, our findings reveal a novel mechanism of targeting a histone demethylase to its functional loci and show the importance of transcription factors in epigenetic regulation.
Results
FYRN and FYRC domains are critical for the biological functions of JMJ14
To analyze whether the FYRN and FYRC domains of JMJ14 are important for its regulation in flowering time and transcriptional regulation, we transformed jmj14-1 mutants with translational fusion constructs consisting of either JMJ14 genomic DNA (JMJ14-HA) or a truncated JMJ14 without FYRN and FYRC domains (JMJ14ΔFYR-HA), each containing an Influenza Hemagglutinin (HA) epitope tags and driven by the native JMJ14 promoter. We selected two transgenic lines from each constructs for further analysis, in which the expression levels of JMJ14 were similar to that in wild-type Columbia (Col; Supplementary Figure S1). JMJ14-HA plants could rescue the early-flowering phenotype of jmj14, whereas the JMJ14ΔFYR-HA plants could not (Figure 1a). Moreover, the expression level of FT in these lines was also in line with their flowering time (Figure 1b).
The FYR (FYRN+FYRC) domain is important for the biological function of JMJ14. (a) The flowering times of Columbia (Col), jmj14-1 and different JMJ14 (Jumonji C (JmjC) domain-containing protein 14) complementary lines under long day condition (16 h light, 8 h dark) at 23 °C. Flowering time was assessed by counting the number of rosette and cauline leaves when the plants flowered. (b) The expression level of flowering locus T (FT) in different JMJ14 complementary lines compared with Col and jmj14-1. (c) The JAP/jmj14-1 homozygous plants were crossed with jmj14-1 and different JMJ14 complementary lines. The leaf phenotypes of F1 plants were observed. The JAP+/- plants under Col background were used as controls. JMJ14-HA and JMJ14ΔFYR-HA indicates the PJMJ14::JMJ14-HA jmj14-1 and PJMJ14::JMJ14ΔFYR-HA jmj14-1 transgenic lines, respectively.
Jawohl:AtSuc2:PDS (JAP) plants contain a transgene that triggers inverted repeat post-transcriptional gene silencing of the endogenous PDS gene and show a JMJ14-dependent photobleaching phenotype [12]. We crossed different JMJ14 transgenic lines to JAP jmj14-1, in which the photobleaching phenotype of JAP plants was suppressed by JMJ14 mutation. The result showed that JMJ14-HA but not JMJ14ΔFYR-HA can derepress the photobleaching phenotype of jmj14-1 (Figure 1c). Taken together, these evidence demonstrate that FYRN and FYRC domains are critical for JMJ14 in regulation of flowering time and transgene silencing.
FYRN and FYRC domains are required for genome-wide JMJ14 targeting
To gain a broader understanding of how FYRN and FYRC domains affect JMJ14 function, we performed chromatin immunoprecipitation (ChIP) followed by sequencing (ChIP-seq) with anti-H3K4me3 antibody in jmj14-1, JMJ14-HA jmj14, JMJ14ΔFYR-HA jmj14, and wild-type Col. We identified 221 genes in jmj14 and 262 genes in JMJ14ΔFYR-HA jmj14 that showed H3K4me3 hypermethylation, among which 177 genes are common between the two data sets (Figure 2a). This indicates that JMJ14ΔFYR-HA cannot rescue the H3K4me3 hypermethylation phenotype in the jmj14 mutant, while H3K4me3 levels of these 177 genes were completely recovered in JMJ14-HA jmj14 to wild-type Col level (Figure 2b). Interestingly, JMJ14ΔFYR-HA showed normal H3K4me3 demethylation activity when overexpressed (Supplementary Figure S2). These results suggest that FYRN and FYRC domains may be essential for recruiting JMJ14 to its endogenous target genes as opposed to being required for enzymatic activity, per se.
JMJ14 (Jumonji C (JmjC) domain-containing protein 14) lost its targets on chromosome in the absent of FYR (FYRN+FYRC) domain. (a) The target genes with twofold increase of H3K4me3 in JMJ14ΔFYR-HA jmj14-1 transgenic plants are significantly overlapped with those in jmj14-1. (b) The H3K4me3 pattern in Columbia (Col), JMJ14-HA jmj14-1, JMJ14ΔFYR-HA jmj14-1, and jmj14-1 plants. TSS, Mid and TTS refers to transcription start site, middle of gene, and transcription termination site, respectively. Genes used for analysis were the common 177 hypermethylated genes of jmj14-1 and JMJ14ΔFYR-HA jmj14-1. (c) The JMJ14 binding level on the direct target genes of JMJ14 in JMJ14-HA jmj14-1, JMJ14ΔFYR-HA jmj14-1, and jmj14-1 plants. The tag counts were normalized in each bin according to the total number of reads. (d) The anti-HA and anti-H3K4me3 chromatin immunoprecipitation sequencing (ChIP-seq) results for At1g21290 locus on genome browser. (e) The anti-HA ChIP-qPCR validation for At1g21290 locus. (f) The anti-H3K4me3 ChIP-qPCR validation for At1g21290 locus. (g) Detection of At1g21290 transcripts by RT-quantitative PCR. Two independent lines of each transgenic plants were used in the qPCR. R1 and R2 indicate the 5′ and 3′ regions of the gene showed in d.
To further determine whether the FYRN and FYRC domains mediate the global targeting of JMJ14, we performed ChIP-seq with anti-HA in both the JMJ14-HA jmj14 and JMJ14ΔFYR-HA jmj14 lines. By analyzing the ChIP-seq results in JMJ14-HA jmj14 transgenic plants, we identified 761 target genes of JMJ14. Strikingly, the binding signals on JMJ14 target genes were completely lost in JMJ14ΔFYR-HA jmj14, which is reminiscent of jmj14 mutant (Figure 2c). Altogether, these results show that FYRN and FYRC domains are essential for proper targeting of JMJ14 to chromatin.
In Arabidopsis, genome-wide distribution patterns of H3K4me3 are found to be exclusively genic and predominantly localized at transcriptional starting sites [10]. Interestingly, we found that JMJ14 occupied the whole gene bodies in a pattern strikingly different from that of H3K4me3 in all the target genes. When more transgenic lines were used for ChIP-quantitative PCR (qPCR) analysis, all target genes tested were validated (Supplementary Figure S3). For example, At1g21290, which codes a copia-like retrotransposon, is one of the JMJ14 targets (Figures 2d and e). In wild-type and JMJ14-HA jmj14-1 plants, the H3K4me3 at At1g21290 was very low, indicating that this transposable element was silenced in normal condition (Figures 2d and f). Hypermethylation of H3K4me3 coupled with increase of gene expression was observed in both jmj14-1 and JMJ14ΔFYR-HA jmj14-1 plants compared with that in Col and JMJ14-HA jmj14-1 plants (Figures 2d and g). These results imply that FYRN and FYRC domains are essential for JMJ14 recruitment to remove H3K4me3 and repress transcription.
JMJ14 interacts with NAC050 and NAC052 through its FYRN and FYRC domains
To identify potential interacting proteins of JMJ14, a yeast two-hybrid screening was performed using the full-length JMJ14 as the bait. From this screening, we identified a pair of putative transcription factors, NAC050 and NAC052 as the top hits. Independently, we created a 3× flag tagged version of JMJ14 driven by its endogenous promoter, and used this line to perform immunoprecipitation followed by mass spectrometry. NAC050 and NAC052 were among the most abundant interacting proteins (Supplementary Table S1). NAC050 and NAC052 belong to a NAC (NAM, ATAF, CUC) transcription factor superfamily, which is specific to plants. There are >100 predicted members of NAC family containing the conserved NAC domain in Arabidopsis [27, 28]. NAC050 and NAC052 are encoded by tandem genes At3g10480 and At3g10490, respectively. Sequence alignment revealed that NAC050 and NAC052 share high homology, with 88% identity (Supplementary Figure S4). In order to dissect the nature of the interaction, we co-expressed full-length coding sequence (CDS) of NAC050 or NAC052 fused with GAL4-AD and JMJ14-GAL4-BD into yeast AH109 strains, and found these combinations activated the ADE2, HIS3, and lacZ reporter genes, further confirming our previous results (Figures 3a and b). To determine which part of JMJ14 mediates the interaction between JMJ14 and NAC050/NAC052, we generated a series of constructs with deletions of different domains of JMJ14, including JMJ14ΔJmjN, JmjN, JMJ14ΔJmjC, JmjC, JMJ14ΔZnF, ZnF, JMJ14ΔFYR, and FYR-only fused with GAL4-BD, respectively (Figure 3b and Supplementary Figure S5A). We found that the interaction between JMJ14 and NAC050/NAC052 was completely abolished after deletion of FYRN and FYRC domains (Figure 3b), whereas deletion of other domains had little effect (Supplementary Figure S5B). We further proved that the FYRN and FYRC domains were sufficient to mediate the interaction between JMJ14 and NAC050/NAC052 in yeast (Figure 3b), indicating that the FYRN and FYRC domains are responsible for the interaction. Interestingly, we found that only FYRN or FYRC domain alone could not interact with NAC050 and NAC052 (Supplementary Figure S5B), suggesting that FYRN and FYRC domains function as an integral. To further verify this interaction, we performed pull-down assays using GST (glutathione S-transferase)-tagged JMJ14 FYRN and FYRC domains as ‘bait’ and maltose binding protein-His-tagged NAC050 or NAC052 as ‘prey’. GST pull-down and immunoblotting assays showed that FYRN and FYRC domains of JMJ14 indeed interact directly with NAC050 and NAC052 in vitro (Figure 3c).
JMJ14 (Jumonji C (JmjC) domain-containing protein 14) interacts with NAC050 and NAC052 through its FYR (FYRN+FYRC) domain in vitro and in vivo. (a) The schematic representation of constructs used for yeast two hybrids. (b) The full-length JMJ14 interacts with NAC050 and NAC052 in yeast (left). Deletion of FYR domain completely abolishes the interaction between JMJ14 and NAC050/NAC052 while FYR domain alone could interact with NAC050/NAC052 in yeast (right). ‘-2’ indicates SD/-Trp-Leu medium, ‘-4’ indicates SD/-Trp-Leu-His-Ade medium with X-α-Gal. (c) Recombinant His-maltose binding protein-NAC050/052 interact with glutathione S-transferase (GST)-FYR directly but not GST in vitro. (d) JMJ14, but not JMJ14ΔFYR, was immunoprecipitated by anti-NAC detected by western blot (WB). JMJ14-HA refers to the PJMJ14::JMJ14-HA jmj14-1 transgenic lines and JMJ14ΔFYR-HA refers to the PJMJ14:: JMJ14ΔFYR-HA jmj14-1 transgenic lines. The red star and the red cross indicate JMJ14-HA and JMJ14ΔFYR-HA fusion proteins, respectively, detected by anti-HA.
In Arabidopsis, both NAC050 and NAC052 localized to the nucleus, as with JMJ14 (Supplementary Figure S6A). Moreover, these two proteins and JMJ14 also shared similar expression pattern by promoter::GUS analysis (Supplementary Figure S6B–D), suggesting that JMJ14 and NAC050/052 may act together in vivo. We further confirmed the interaction in vivo by co-immunoprecipitation assays. Antibodies that could recognize both NAC050 and NAC052 were used to immunoprecipitate proteins from lysate of 12-day-old seedlings containing either JMJ14-HA or JMJ14ΔFYR-HA, respectively. Only the full-length JMJ14-HA could be immunoprecipitated but not the truncated JMJ14 without FYRN and FYRC domains (JMJ14ΔFYR-HA; Figure 3d), confirming that JMJ14 interacts with NAC050/052 in Arabidopsis and such interaction is dependent on the FYRN and FYRC domains.
NAC050 and NAC052 share widespread common targets with JMJ14 in direct binding and gene regulation
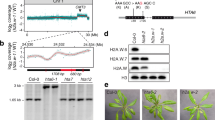
NAC family proteins have been shown to have important roles in almost every aspects of plant growth and development, including vegetative and floral development, root formation, auxin signaling, and biotic and abiotic stress response [29]. To determine the biological functions of NAC050 and NAC052 and their relationship with JMJ14, we identified the T-DNA insertion mutants from ABRC and WiscDsLox T-DNA Collection (Figure 4a), in which nac050-1, nac050-2, and nac052-2 are null alleles, whereas nac052-1 represents a weak allele with partial reduction of full-length cDNA (Figure 4b).
NAC050 and NAC052 share common targets in direct binding with JMJ14 (Jumonji C (JmjC) domain-containing protein 14). (a) Gene structure of NAC050 and NAC052 and the T-DNA insertion sites of nac050 and nac052 mutants. Black bars, gray bars and black lines indicate coding exons, UTRs and introns, respectively. T-DNA insertions are indicated by triangles. Scale bar, 500 bp. (b) Detection of full-length CDS of NAC050 and NAC052 in the nac050 and nac052 mutants. Actin was used as an internal control. (c) The targets of NAC050 and NAC052, identified by chromatin immunoprecipitation sequencing (ChIP-seq), are significantly overlapped. (d) Nearly half of JMJ14’s targets are co-localized by both NAC050 and NAC052. (e) Three target genes showed co-localized ChIP-seq signal of JMJ14, NAC050, and NAC052 on genome browser. JMJ14-HA, NAC050-HA, and NAC052-HA indicates the PJMJ14::JMJ14-HA jmj14-1, PNAC050::NAC050-HA nac050-1, and PNAC052::NAC052-HA nac052-2 transgenic lines, respectively.
As NAC050 and NAC052 interact with JMJ14, we hypothesized that NAC050 and NAC052 may have common targets with JMJ14 in the genome. We therefore performed ChIP-seq analysis using anti-HA antibody in PNAC050::NAC050-HA nac050-1 and PNAC052::NAC052-HA nac052-2, respectively. The wild-type Col plants that do not contain HA tags were used as a negative control. We identified 721 and 787 target genes of NAC050 and NAC052, respectively, and more than three quarters (597) are common targets (Figure 4c), among which, >60% (369 out of 597) are also bound by JMJ14 (Figure 4d). Moreover, the binding signals of NAC050 and NAC052 distributed across the whole gene body of their targets as a similar pattern as that of JMJ14 (Supplementary Figure S7). The common targets of JMJ14, NAC050, and NAC052 were predominantly enriched in negative regulation of molecular function and protein phosphorylation in terms of biological processes (Supplementary Figure S8), indicating that they may be involved in signal transduction. For example, the binding signals of JMJ14, NAC050, and NAC052 exhibit significant overlap on target genes At1g72460 and At2g32510, which encode two kinase proteins; the previously identified transposable elements At1g21290 showed a similar pattern as well (Figure 4e). The significant overlapping of target genes suggests that NAC050 and NAC052 may function redundantly and act together with JMJ14 to collaboratively regulate a large number of genes in Arabidopsis.
NAC050 and NAC052 bind a specific palindromic DNA motif
Previous reports showed that NAC transcription factors can be recruited by cis-elements on their target genes [30, 31]. Therefore, we speculated that NAC050 and NAC052 could also bind specific DNA sequences. To identify the DNA motif that was bound by NAC050 and NAC052, we analyzed the DNA sequences around the binding peaks of NAC050 and NAC052 within the genome using MEME-ChIP programs [32]. A palindromic sequence CTTGNNNNNCAAG intensively exists in both NAC050 and NAC052 binding peaks (Figure 5a), which is also enriched in JMJ14 binding peaks (Figure 5a). To determine the binding ability of NAC050 and NAC052 in vitro, we synthesized a 30-bp DNA probe that contains the CTTGNNNNNCAAG core sequence in At1g72460, one of the common targets of NAC050, NAC052, and JMJ14, for electrophoretic mobility shift assays. In contrast to GST-FYR and GST alone, which did not bind this sequence, both NAC050 and NAC052 proteins displayed strong binding affinity to this sequence (Figure 5b). To further verify the specificity of the binding motifs, we generated a series of mutated version of the probe (P) (Supplementary Figure S9A) and performed electrophoretic mobility shift assay. We found that mutation of either CTTG and/or CAAG core sequences reduced the binding significantly (Supplementary Figure S9B). In addition, we found that the DNA sequences rather than the inverted repeated signature is important for the binding (Supplementary Figures S9A and B). We also found that the linker length between CTTG and CAAG is also very critical for NAC050 and NAC052 binding (Supplementary Figures S9A and B). These findings suggest that NAC050 and NAC052 could directly bind the ‘CTTGNNNNNCAAG’ motif, which may be necessary to facilitate the recruitment of JMJ14 to its targets.
Identification of the DNA motif bound by NAC050 and NAC052. (a) A palindromic DNA motif CTTGNNNNNCAAG identified in the binding regions of NAC050, NAC052, and JMJ14. (b) The binding assay using different amount of GST (glutathione S-transferase)-fused recombinant proteins showed significant binding affinity of NAC050 and NAC052 but not FYR (FYRN+FYRC) domain of JMJ14 (Jumonji C (JmjC) domain-containing protein 14) to the probe.
NAC052 and NAC050 are involved in RNA silencing
To further test whether NAC050 and NAC052 function with JMJ14, we crossed nac050 and nac052 with JAP lines. In parallel with the genetic relation between jmj14 and JAP lines, mutation of NAC052, and to a lesser extent NAC050, indeed suppress the photobleaching phenotype of JAP (Figure 6a). Consistent with reduced photobleaching, JAPnac052-2 and JAPnac050-2 also displayed lower levels of PDS small interfering RNAs (Figure 6b). All these phenotypes are similar to those seen in jmj14 mutant. To further confirm this observation, we then crossed two PNAC052::NAC052-HA nac052-2 lines into JAPnac052-2 and found that NAC052-HA could fully rescue the phenotype of JAPnac052-2 (Figure 6c). These results confirmed that the loss of photobleaching phenotype in JAPnac052 was caused by the mutation of NAC052. Thus, we concluded that NAC050 and NAC052 cooperate with JMJ14 to positively regulate RNA silencing.
Photobleaching suppression of nac052 and to a less extent nac050 in transgenic lines. (a) The leaf photobleaching phenotype of nac050 and nac052 compared with jmj14-1 under JAP background. The upper panel shows the 12-day plants and the bottom panel shows the 3-week plants. (b) Detection of PDS small interfering RNAs (siRNAs) in JAP lines under different mutant background. (c) The JAP/nac052-2 homozygous plants were crossed with nac052-2 and NAC052 complementary lines. The leaf phenotypes of F1 plants were observed. The JAP+/- plants under Columbia (Col) background were used as controls. NAC052-HA refers to the PNAC052::NAC052-HA nac052-2 transgenic lines. (d) The model for JMJ14 targeting mediated by NAC050/052.
Discussion
JmjC-domain-containing proteins have been found to have important roles in growth and development in both animals and plants. However, how these proteins are recruited to their target genes is poorly understood. In animals, two modes were reported for chromatin-associated protein targeting: namely mediated by a specific domain of such protein or coordinated with other chromatin-associated proteins. For example, two H3K36 histone demethylases, lysine demethylase 2A (KDM2A) and KDM2B [33, 34], are recruited to non-methylated CpG islands by their zinc-finger CxxC (ZF-CxxC) domain [35–38]. Jarid1a, one of the H3K4 demethylases in KDM5 subgroup, is recruited to different target genes by associating with polycomb repressive complex 2, a histone H3K27 methyltransferase containing complex [39, 40], or G9a, a histone H3K9 methyltransferase [41, 42].
JMJ14 belongs to KDM5/JARID1 subfamily and possesses H3K4 demethylase activity [9–11, 17]. It has been shown to be involved in transgene silencing and flowering time regulation [9–12, 16]. In this work, we demonstrate that the FYRN and FYRC domains of JMJ14 have crucial roles in transcriptional gene silencing and flowering time regulation. Deletion of FYRN and FYRC domains causes mistargeting of JMJ14 over several hundred of genes and H3K4me3 hypermethylation mimicking jmj14 mutants.
We also reveal that JMJ14 interacts with two putative transcription factors, NAC050 and NAC052, through its FYRN and FYRC domains. Although FYRN and FYRC were identified as two domains separately, our results showed these two domains function only when they are both presence. This is consistent with the previous structural evidence that FYRN and FYRC form a single domain with an α+β fold [23]. By genome-wide analysis, we identified a significant palindromic DNA motif CTTGNNNNNCAAG in JMJ14, NAC050, and NAC052 binding regions. We prove that NAC050 and NAC052 but not FYRN and FYRC domains of JMJ14 bind to this DNA motif directly. Therefore, we propose that NAC050 and NAC052 bind to their targets by recognizing the palindromic DNA motifs and recruit JMJ14 through the interaction with FYRN and FYRC domains (Figure 6d). Besides, the overlapped binding peaks of JMJ14 and NAC050/052, we also calculated the density of the CTTGNNNNNCAAG motif in the binding regions of JMJ14 alone and the common binding regions of NAC050 and NAC052. According to our analysis, the motif is still dominantly enriched in the non-overlapped regions, which is present in ~85% of the non-overlapped JMJ14 binding regions and 73% of non-overlapped NAC050/052 binding regions (Supplementary Figure S9C). This result may suggest that NAC050/052 first formed a stable interaction by motif matching before recruitment of JMJ14 to the targeted genome, and then left after JMJ14 is ready for action.
Mutations in NAC050 and NAC052 derepress transgene silencing similar with that of jmj14, which further prove that NAC050/052 act together with JMJ14 in vivo. We also noticed that JAPnac052 showed more reduced photobleaching phenotype than that of JAPnac050. This is consistent with more reduction of PDS small interfering RNA accumulation in JAPnac052 than that in JAPnac050, which suggest that NAC052 has predominant role and such level of reduction is sufficient to suppress the photobleaching phenotype. Therefore, we uncovered a novel targeting mechanism that uncharacterized domains of JMJ14 associate with a pair of NAC transcription factors to promote transcriptional repression. Recently, it has been shown that a pair of transcription factors Pax3 and Pax9 bind to satellite repeats sequences and recruit H3K9 methyltransferase Suv39h to establish H3K9me3 and silence the heterochromatin in mouse [43], suggesting a conserved function of transcription factors in silencing in both animals and plants.
In addition, Wang et al. [44] showed that JMJ14 acts together with polycomb repressive complex 1-like complex that is responsible for the maintenance of H3K27me3. In Arabidopsis, H3K4me3 and H3K27me3 are known to be mutually exclusive. Our finding that JMJ14 recruitment by NAC050 and NAC052 to specific sequences leads to removal of H3K4me3 suggests that JMJ14 is the key regulator in establishing H3K27me3 repressive domain in some of the Polycomb targets. It is interesting to dissect the role of JMJ14 in regulating Polycomb-mediated gene silencing at genome-scale in the future.
Materials and Methods
Plant materials
All Arabidopsis plants were grown at 23 °C under long day condition (16 h light, 8 h dark). JAP transgenic lines were obtained from Dr David Baulcombe. The mutants alleles used are as follows: jmj14-1(SALK_135712), nac050-1 (SAIL_841_F01), nac050-2 (SALK_026244C), nac052-1 (SALK_056304C), and nac052-2 (WiscDsLoxHs027_03D).
Yeast two-hybrid assays
For yeast two-hybrid screening, full-length CDS of JMJ14 was cloned into pGBKT7 vector using primers CX5024 and CX5025. A mixture of RNAs from 10-day-old seedlings and flowers of Arabidopsis were used to construct the cDNA library. Library construction and screening were performed with Matchmaker Library Construction & Screening Kits (Clontech 630445, Japan). The full-length CDS of NAC050 and NAC052 were cloned into pGADT7 vector using primers CX8980 and CX8981 (for NAC050) or CX8984 and CX8985 (for NAC052). The truncated JMJ14 with different domain deletions were cloned using a QuickChange kit (Stratagene, Santa Clara, CA, USA) with primers CX8988 and CX8989 (for JMJ14ΔJmjN), CX8990 and CX8991 (for JMJ14ΔJmjC), CX8992 and CX8993 (for JMJ14ΔZnF), or CX8994 and CX8995 (for JMJ14ΔFYR). The different domains of JMJ14 were cloned using primers HX2032 and HX2033 (for JmjN), HX2034 and HX2035 (for JmjC), HX2036 and HX2037 (for ZnF), HX2038 and HX2041 (for FYR), HX2038 and HX2039 (for FYRN), and HX2040 and HX2041 (for FYRC). The bait and prey vectors were cotransformed into yeast AH109 strain following the manufacturer’s handbook (Clontech Yeast Protocols Handbook). The cotransformed yeast clones were first cultured in SD/-Leu/-Trp liquid medium. The same amount of yeast in liquid medium was plated on SD/-Leu/-Trp and SD/-Ade/-His/-Leu/-Trp medium containing 20 μg ml−1 X-α-Gal respectively. Primer sequences can be found in Supplementary Table S2.
GST pull-down
The full-length CDS of NAC050 and NAC052 were amplified with primers HX2046 and CX9579 (for NAC050) or HX2047 and CX9581 (for NAC052) and inserted into the expression vector pMAL-C2-maltose binding protein (XF510). The FYR domain of JMJ14 was amplified with primers HX0406 and CX5012 and inserted into the expression vector pMAL-C2-GST (XF760). The fragments were inserted into pMAL-C2-maltose binding protein and pMAL-C2-GST by a ligation-independent cloning method as previously described [45]. All the recombinant proteins were expressed and purified from E scherichia coli strain BL21 RIL(BL21 CP, Stratagene). For pull-down assays, bait proteins were bound to glutathione beads and blocked overnight with 5% bovine serum albumin in pull-down buffer (50 mM Tris pH 7.5, 300 mM NaCl, and 0.1% CA-630). Then the complexes were incubated with target proteins for 45 min, washed eight times with pull-down buffer. Eluates were analyzed by western blotting using anti-His antibodies (Abmart M20001, Shanghai, China). Primer sequences can be found in Supplementary Table S2.
Co-immunoprecipitation assay
One gram of 12-day-old seedlings of Arabidopsis were ground to powder in liquid N2 and suspended in 1 ml of PEN-140 buffer (140 mM NaCl, 2.7 mM KCl, 25 mM Na2HPO4, 1.5 mM KH2PO4, 0.01 mM EDTA, and 0.05% CA-630). The supernatant was incubated 2 h with 40 μl Dynabeads Protein G (Invitrogen 10004D, Waltham, MA, USA) conjugated with antibodies that could recognize both NAC050 and NAC052 (Supplementary Figure S10). The beads were washed five times with PEN-400 buffer (400 mM NaCl, 2.7 mM KCl, 25 mM Na2HPO4, 1.5 mM KH2PO4, 0.01 mM EDTA, and 0.05% CA-630) and then analyzed by western blotting using anti-HA antibodies (Sigma H6908, St Louis, MO, USA).
ChIP assay
ChIP assays were performed using 1 g of 12-day-old seedlings as previously described with minor modifications [10]. For anti-HA ChIP, chromatin was fragmented by micrococcal nuclease (NEB M0247S) in the micrococcal nuclease digestion buffer (50 mM Tris pH 8.0, 0.32 M sucrose, 4 mM MgCl2, and 1 mM CaCl2) instead of sonication before nuclei lysis. Antibodies used in ChIP were anti-H3K4me3 (Millipore 07-473, Darmstadt, Germany), anti-H3 (Abcam ab1791, Cambridge, MA, USA) and anti-HA (Sigma H6908). Primers for ChIP-qPCR can be found in Supplementary Table S2.
ChIP-seq analysis
ChIPed DNA was ligated with Illumina single-end genome sequencing adapters, size fractionated to obtain 300-bp fragments, PCR amplified and sequenced according to standard protocols (single-end 36 cycles). The reads were aligned to Arabidopsis thaliana genome build TAIR10 by Bowtie 2 [46] using default parameters with local alignment model. Duplicated reads and low-mapping quality reads were identified and removed with SAMtools [47]. Enriched intervals were identified by MACS [48] with default parameters. Density maps of reads for visualization were calculated by SAMtools as well as programs in University of California, Santa Cruz, genome browser based on counts of the 200-bp extension of sequencing reads in the 3ʹ direction (as described previously in Ernst et al. [49]) after total reads normalization.
Electrophoretic mobility shift assay
The full-length CDS of NAC050 and NAC052 were amplified with primers HX2046 and CX9579 (for NAC050) or HX2047 and CX9581 (for NAC052). The FYR domain of JMJ14 was amplified with primers HX0406 and CX5012. The fragments were inserted into the expression vector pMAL-C2-GST (XF760) by a ligation-independent-cloning method as previously described [45]. All the recombinant proteins were expressed and purified from E. coli strain BL21 RIL (BL21 CP, Stratagene). Ten or 50 ng of purified proteins were incubated with 0.3 pM of each 32P-labeled double-stranded DNA oligos in binding buffer (25 mM Tris-HCl, 100 mM NaCl, 2.5 mM MgCl2, 0.1% CA-630, 10% glycerol, and 0.5 mM Dithiothreitol) on ice for 1 h. The binding reaction mixture was loaded onto a 6% non-denaturing polyacrylamide gel at 80 V for 1 h. The DNA was detected by the radioactive signal. Primer sequences can be found in Supplementary Table S2. The probes sequences are listed in Supplementary Figure S9A.
Transgene
The full-length JMJ14 genomic fragment with 1.5 kb of promoter was amplified using primers CX5033 and CX3599. The truncated JMJ14 without FYRN and FYRC (FYR) domains was amplified using primers CX5033 and CX7385. The fragments were then cloned into pENTR/D-TOPO (Invitrogen) and introduced by LR reaction into pEarleyGate301 (pEG301) vector with a C-terminal HA tag (PJMJ14::JMJ14-HA and PJMJ14::JMJ14ΔFYR-HA) [50]. The full-length CDS of NAC050 and NAC052 were introduced into pEarleyGate102 (pEG102) vector with a C-terminal CFP-HA tag (P35S::NAC050-CFP-HA and P35S::NAC052-CFP-HA) using primers CX8188 and CX8189 (for NAC050) or CX8191 and CX8194 (for NAC052). P35S::JMJ14-YFP-HA was constructed as previously described [10]. For GUS staining vectors, ~0.8 kb of 3ʹ untranslated regions of JMJ14, NAC050, and NAC052 were cloned and introduced into pCambia1300-GFPGUS (XF1764) after NcoI digestion. The full-length genomic regions with native promoter of JMJ14, NAC050, and NAC052 were then amplified and constructed into pCambia1300-GFPGUS-UTR. The following primers are used: CX9272 and CX9273 for NAC050 genomic region, CX9274 and CX9275 for NAC050 3ʹUTR, CX9276 and CX9277 for NAC052 genomic region, CX9278 and CX9279 for NAC052 3ʹUTR, CX9280 and CX9281 for JMJ14 genomic region, and CX9282 and CX9283 for JMJ14 3ʹUTR. For NAC050 and NAC052 complementary vectors, ~0.8 kb of 3ʹUTR of NAC050 and NAC052 were cloned and introduced into pCambia1300-HA (XF0791) after PstI digestion. The full-length genomic sequences with native promoter of NAC050 and NAC052 were then amplified and constructed into pCambia1300-HA-UTR by EcoRI and BamHI double digestion. The primers used are as follows: CX9272 and CX9273 for NAC050 genomic region, HX2078 and HX2079 for NAC050 3ʹUTR, CX9276 and CX9277 for NAC052 genomic region, and HX2080 and HX2081 for NAC052 3ʹUTR. The constructs were introduced into A grobacterium tumefaciens cells (strain EHA105) and then stably transformed into Arabidopsis using the floral dip method [51]. Primer sequences can be found in Supplementary Table S2.
Flowering time assessment
All plants were grown in soil side by side at 23 °C under long day (16 h light and 8 h dark) conditions. Flowering time was assessed by counting the number of rosette and cauline leaves when the plants flowered. At least 15 plants were counted for each line.
Affinity purification and mass spectrometry
Purification of JMJ14-3×Flag was performed as described in Law et al. [52] with minor modifications. In brief, 15 g of flower tissue collected from transgenic T4 plants, or from Col plants as a negative control, were ground with a mortar and pestle in liquid nitrogen and suspended in 45 ml of lysis buffer (50 mM Tris (pH 7.6), 150 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.1% NP-40, 0.5 mM DTT, 1 mg ml−1 pepstatin, 1 mM Phenylmethylsulfonyl fluoride, and 1 protease inhibitor cocktail tablet (Roche, 14696200, Basel, Switzerland)). Following centrifugation to remove insoluble plant debris, lysate was incubated with 125 μl of Dynabeads (M-270 Epoxy, Invitrogen, 143.01) that had been conjugated with Flag antibody (Sigma F 3165) according to the manufacturer’s instructions. After incubation at 4 °C with rotation for 2.5 h, the Flag beads were washed seven times with lysis buffer. Proteins were then eluted from the Flag beads by competition with 150 μl of 100 μg ml−1 of 3×Flag peptide (Sigma, F 4799) five times at room temperature. Mass spectrometry and analysis was performed exactly as described in Law et al. [52].
RNA gel blot and transcription level analysis
Total RNA was extracted from 12-day-old seedlings using TRNzol reagent (Tiangen, Beijing, China). RNAs (15 μg per lane) were separated in an agarose gel containing 1% formaldehyde, blotted onto Hybond N+ membrane (GE Healthcare RPN119B, Little Chalfont, UK), and probed with JMJ14 cDNA, which was amplified using primers CX5024 and CX5025. PDS small interfering RNA gel blot was performed exactly as previous report [53]. Reverse transcription reactions were performed using TransScript II First-Strand cDNA Synthesis SuperMIX (TransGen AH301-02, Beijing, China). qPCR was performed using a CFX96 Real-time PCR instrument (Bio-Rad, Hercules, CA, USA) with RealSYBR Mixture (CWBIO and CW0760). UBC (At5g25760) was used as the internal control of Reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Primers for RT-qPCR or RT-PCR are listed in Supplementary Table S2.
Histochemical GUS staining
Histochemical GUS staining was performed according to the standard procedure [54]. Twelve-day-old seedlings of pCambia1300-GFP-GUS transgenic lines were incubated in GUS staining buffer (50 mM sodium phosphate buffer (pH 7.2), 10 mM EDTA, 0.2% Triton X-100, and 2 mM potassium ferrocyanide/ferricyanide) with 2 mM X-glucuronide at 37 °C. After staining, the samples were cleared and stored in 70% ethanol for microscopic analysis.
In vivo histone demethylation assay
The truncated JMJ14 without FYRN and FYRC (FYR) domains was amplified using primers CX3598 and CX7385. The fragments were then cloned into pENTR/D-TOPO (Invitrogen) and introduced by LR reaction into pEarleyGate101 (pEG101) vector with a C-terminal YFP-HA tag (P35S::JMJ14ΔFYR-YFP-HA). Tobacco (Nicotiana benthamiana) leaves was infiltrated with A. tumefaciens EHA105 strains containing P35S::JMJ14ΔFYR-YFP-HA vector to overexpress JMJ14ΔFYR-YFP-HA. The demethylation assay was carried out as previously described [10]. Immunolabeling was performed using histone methylation-specific antibodies (H3K4me3: Millipore 07-473, 1:100; H3K4me2: Millipore 07-030, 1:500; and H3K4me1: Millipore 07-436, 1:100). At least 25 pairs of transfected or non-transfected nuclei were observed in the same field of view. Quantification was done using ImageJ (National Institutes of Health, Bethesda, MD, USA) software by comparing Alexa Fluor 555-staining density of nuclei overexpressing JMJ14ΔFYR-YFP-HA to that of the local neighboring wild-type nuclei.
Data deposition
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus [55] and are accessible through GEO Series accession number GSE60084 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE60084).
References
Martin C, Zhang Y . The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 2005; 6: 838–849.
Liu C, Lu F, Cui X, Cao X . Histone methylation in higher plants. Annu Rev Plant Biol 2010; 61: 395–420.
Mosammaparast N, Shi Y . Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem 2010; 79: 155–179.
Black JC, Van Rechem C, Whetstine JR . Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell 2012; 48: 491–507.
Noh B, Lee SH, Kim HJ, et al. Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 2004; 16: 2601–2613.
Saze H, Shiraishi A, Miura A, Kakutani T . Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science 2008; 319: 462–465.
Yu X, Li L, Li L, Guo M, Chory J, Yin Y . Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc Natl Acad Sci USA 2008; 105: 7618–7623.
Miura A, Nakamura M, Inagaki S, Kobayashi A, Saze H, Kakutani T . An Arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. EMBO J 2009; 28: 1078–1086.
Jeong JH, Song HR, Ko JH, et al. Repression of FLOWERING LOCUS T chromatin by functionally redundant histone H3 lysine 4 demethylases in Arabidopsis. PloS One 2009; 4: e8033.
Lu F, Cui X, Zhang S, Liu C, Cao X . JMJ14 is an H3K4 demethylase regulating flowering time in Arabidopsis. Cell Res 2010; 20: 387–390.
Yang W, Jiang D, Jiang J, He Y . A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. Plant J 2010; 62: 663–673.
Searle IR, Pontes O, Melnyk CW, Smith LM, Baulcombe DC . JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes Dev 2010; 24: 986–991.
Inagaki S, Miura-Kamio A, Nakamura Y, et al. Autocatalytic differentiation of epigenetic modifications within the Arabidopsis genome. EMBO J 2010; 29: 3496–3506.
Deleris A, Greenberg MV, Ausin I, et al. Involvement of a Jumonji-C domain-containing histone demethylase in DRM2-mediated maintenance of DNA methylation. EMBO Rep 2010; 11: 950–955.
Lu F, Cui X, Zhang S, Jenuwein T, Cao X . Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet 2011; 43: 715–719.
Le Masson I, Jauvion V, Bouteiller N, Rivard M, Elmayan T, Vaucheret H . Mutations in the Arabidopsis H3K4me2/3 demethylase JMJ14 suppress posttranscriptional gene silencing by decreasing transgene transcription. Plant Cell 2012; 24: 3603–3612.
Lu F, Li G, Cui X, Liu C, Wang XJ, Cao X . Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J Integr Plant Biol 2008; 50: 886–896.
Iwase S, Lan F, Bayliss P, et al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 2007; 128: 1077–1088.
Li F, Huarte M, Zaratiegui M, et al. Lid2 is required for coordinating H3K4 and H3K9 methylation of heterochromatin and euchromatin. Cell 2008; 135: 272–283.
Yang H, Han Z, Cao Y, et al. A companion cell-dominant and developmentally regulated H3K4 demethylase controls flowering time in Arabidopsis via the repression of FLC expression. PLoS Genet 2012; 8: e1002664.
Daser A, Rabbitts TH . Extending the repertoire of the mixed-lineage leukemia gene MLL in leukemogenesis. Genes Dev 2004; 18: 965–974.
Finn RD, Mistry J, Schuster-Bockler B, et al. Pfam: clans, web tools and services. Nucleic Acids Res 2006; 34: D247–D251.
Garcia-Alai MM, Allen MD, Joerger AC, Bycroft M . The structure of the FYR domain of transforming growth factor beta regulator 1. Protein Sci 2010; 19: 1432–1438.
Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, Avramova Z . ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr Biol 2003; 13: 627–637.
Ding Y, Fromm M, Avramova Z . Multiple exposures to drought 'train' transcriptional responses in Arabidopsis. Nat Commun 2012; 3: 740.
Grafi G, Zemach A, Pitto L . Methyl-CpG-binding domain (MBD) proteins in plants. Biochim Biophys Acta 2007; 1769: 287–294.
Ooka H, Satoh K, Doi K, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 2003; 10: 239–247.
Jensen MK, Kjaersgaard T, Nielsen MM, et al. The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem J 2010; 426: 183–196.
Olsen AN, Ernst HA, Leggio LL, Skriver K . NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 2005; 10: 79–87.
Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K . NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta 2012; 1819: 97–103.
Puranik S, Sahu PP, Srivastava PS, Prasad M . NAC proteins: regulation and role in stress tolerance. Trends Plant Sci 2012; 17: 369–381.
Machanick P, Bailey TL . MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 2011; 27: 1696–1697.
Tsukada Y, Fang J, Erdjument-Bromage H, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006; 439: 811–816.
He J, Kallin EM, Tsukada Y, Zhang Y . The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b). Nat Struct Mol Biol 2008; 15: 1169–1175.
Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ . CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell 2010; 38: 179–190.
He J, Shen L, Wan M, Taranova O, Wu H, Zhang Y . Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol 2013; 15: 373–384.
Farcas AM, Blackledge NP, Sudbery I, et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. eLife 2012; 1: e00205.
Wu X, Johansen JV, Helin K . Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol Cell 2013; 49: 1134–1146.
Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K . Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev 2008; 22: 1345–1355.
Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002; 298: 1039–1043.
Chaturvedi CP, Somasundaram B, Singh K, et al. Maintenance of gene silencing by the coordinate action of the H3K9 methyltransferase G9a/KMT1C and the H3K4 demethylase Jarid1a/KDM5A. Proc Natl Acad Sci USA 2012; 109: 18845–18850.
Tachibana M, Sugimoto K, Nozaki M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 2002; 16: 1779–1791.
Bulut-Karslioglu A, Perrera V, Scaranaro M, et al. A transcription factor-based mechanism for mouse heterochromatin formation. Nat Struct Mol Biol 2012; 19: 1023–1030.
Wang Y, Gu X, Yuan W, Schmitz RJ, He Y . Photoperiodic control of the floral transition through a distinct polycomb repressive complex. Dev Cell 2014; 28: 727–736.
Eschenfeldt WH, Lucy S, Millard CS, Joachimiak A, Mark ID . A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol Biol 2009; 498: 105–115.
Langmead B, Salzberg SL . Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9: 357–359.
Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25: 2078–2079.
Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008; 9: R137.
Ernst J, Kheradpour P, Mikkelsen TS, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011; 473: 43–49.
Earley KW, Haag JR, Pontes O, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 2006; 45: 616–629.
Clough SJ, Bent AF . Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 1998; 16: 735–743.
Law JA, Ausin I, Johnson LM, et al. A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr Biol 2010; 20: 951–956.
Smith LM, Pontes O, Searle I, et al. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell 2007; 19: 1507–1521.
Jefferson RA, Kavanagh TA, Bevan MW . GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 1987; 6: 3901–3907.
Edgar R, Domrachev M, Lash AE . Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002; 30: 207–210.
Acknowledgements
We thank the Arabidopsis Biological Resource Center for providing T-DNA insertion lines, Dr David Baulcombe for JAP plants. This work was supported by the National Basic Research Program of China (grant nos. 2011CB915401 to X Cao and 2013CB967300 to Xia Cui), the National Natural Science Foundation of China (grant nos. 31271363 to X Cui and 31330020 to X Cao), Genetically Modified Breeding Major Projects (grant no. 2014ZX08010-002 to X Cao), the State Key Laboratory of Plant Genomics, and the post-doctoral fellowship to (BZ and Xiekui Cui). Work in the Jacobsen laboratory was supported by National Institutes of Health grant GM60398. SEJ is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Zhang, S., Zhou, B., Kang, Y. et al. C-terminal domains of histone demethylase JMJ14 interact with a pair of NAC transcription factors to mediate specific chromatin association. Cell Discov 1, 15003 (2015). https://doi.org/10.1038/celldisc.2015.3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/celldisc.2015.3
This article is cited by
-
Identification of epigenetically regulated genes involved in plant-virus interaction and their role in virus-triggered induced resistance
BMC Plant Biology (2024)
-
Genome-wide identification and expression analysis of the JMJ-C gene family in melon (Cucumis melo L.) reveals their potential role in fruit development
BMC Genomics (2023)
-
Arabidopsis TRB proteins function in H3K4me3 demethylation by recruiting JMJ14
Nature Communications (2023)
-
High Temperature Triggers Differential Expression of JUMONJI C (JmjC) Domain-Containing Histone Demethylase Genes in Leaf and Stolon Tissues of Potato (Solanum tuberosum L.) Genotypes
Journal of Plant Growth Regulation (2023)
-
Contrasting epigenetic control of transgenes and endogenous genes promotes post-transcriptional transgene silencing in Arabidopsis
Nature Communications (2021)