Abstract
The Warburg effect is an important characteristic of tumor cells, making it an attractive therapeutic target. Current anticancer drug development strategies predominantly focus on inhibitors of the specific molecular effectors involved in tumor cell proliferation. These drugs or natural compounds, many of which target the Warburg effect and the underlying mechanisms, still need to be characterized. To elucidate the anticancer effects of a natural diterpenoid, oridonin, we first demonstrated the anticancer activity of oridonin both in vitro and in vivo in colorectal cancer (CRC) cells. Then miRNA profiling of SW480 cells revealed those intracellular signaling related to energy supply was affected by oridonin, suggesting that glucose metabolism is a potential target for CRC therapy. Moreover, our results indicated that oridonin induced metabolic imbalances by significantly inhibiting glucose uptake and reducing lactate export through significantly downregulating the protein levels of GLUT1 and MCT1 in vitro and vivo. However, the ATP level in oridonin-treated CRC cells was not decreased when oridonin blocked the glucose supply, indicating that oridonin induced autophagy process, an important ATP source in cancer cells. The observation was then supported by the results of LC3-II detection and transmission electron microscopy analysis, which confirmed the presence of autophagy. Furthermore, p-AMPK was rapidly deactivated following oridonin treatment, resulting in downregulation of GLUT1 and induction of autophagy in the cancer cells. Thus our finding helped to clarify the anticancer mechanisms of oridonin and suggested it could be applied as a glucose metabolism-targeting agent for cancer treatment.
Similar content being viewed by others
Main
One of the hallmarks of cancer cells is enhanced glucose metabolism and dramatically altered nutrient utilization compared to normal tissues, which was first described by Otto Warburg in the early 1900s1 and is now known as the Warburg effect. Glucose is a major source of energy in cancer cells that generates ATP almost through glycolysis, a far less efficient energy generator than mitochondrial respiration.2 As the survival of cancer cells predominantly depend on the high rate of glucose consumption and elevated glycolysis, targeting the Warburg effect and glucose metabolism has become an important strategy for cancer therapy. Several factors, including mitochondrial defects, oncogenic stimuli, hypoxia, and aberrantly enhanced expression of glycolytic enzymes, are now considered to be important drivers of the Warburg effect.2, 3 Due to their critical role in cancer cells, these glucose metabolism-related proteins and enzymes are believed to be potential targets for drug design and cancer therapy.4
There are two main strategies under investigation that target glucose metabolism. One focuses on the regulation of glycolytic flux-related proteins including Gluts, MCTs, lactate dehydrogenase A (LDHA), HK2, and PKM2.5, 6 This strategy aims to directly regulate the glucose supply and glycolytic pathways to control the energy production in cancer cells. Another strategy focuses on factors believed to be central for metabolic regulation, including HIF-1a, c-Myc, AKT, mTOR, and AMPK.7 These factors control the abundance of proteins that regulate the glucose and energy supply of cancer cells. Several compounds, including WZB117, rapamycin, berberine, and metformin, have been shown to regulate glucose metabolism and exhibit anticancer activities.8, 9, 10, 11 However, the mechanisms of anticancer activities by which natural compounds targeting glucose metabolism still need to investigate.
Oridonin is an active diterpenoid isolated from Rabdosia rubescens in the 1970s that has potent anti-tumor activities in many types of human cancer both in vitro and in vivo.12, 13 Oridonin treatment had multiple effects such as inhibition of proliferation and induction of cell cycle arrest, apoptotic and autophagic pathways in a variety of cancer cells including those of colorectal carcinoma.14, 15, 16, 17 Previous studies have indicated that various factors, including Akt, ERK, FAS, ROS, NF-κB, PI3K, and RTK, were involved in the anticancer activity of oridonin.17, 18, 19 Although various mechanisms were proposed to explain the anticancer activity of oridonin, most of the research has been scattered. Oridonin was reported to suppress t(8;21) acute myeloid leukemic (AML) cells via cleavage of the AML1–ETO fusion protein.20 Our lab reported that oridonin induced c-MYC proteasomal degradation in chronic myeloid leukemia.21 Thus, in contrast to these studies performed in leukemic cells, the underlying mechanisms of oridonin-induced anticancer activity in specific solid tumors and which type of solid tumor shows the best response have not been elucidated, and further studies are needed to provide evidence supporting the use of oridonin as a antineoplastic agent.
To elucidate the mechanism of oridonin in CRC cells, we investigated the miRNA profile by high-throughput sequencing both in the absence and presence of oridonin in CRC cells. These results prompted us to focus on the regulation of the Warburg effect and glucose metabolism. We then confirmed that oridonin affected the cellular glucose supply and lactate exportation via AMPK-related glucose transporter 1 (GLUT1) and monocarboxylate transporter 1 (MCT1) regulation. This mechanism could be a trigger that induces cancer cell autophagy and accelerates cell death. Our findings showed that oridonin could affect glucose metabolism and induce autophagy through a metabolism-related pathway.
Results
Oridonin causes cell death through an atypical apoptosis manner in p53-mutated CRC cells and exhibits anticancer activity both in vitro and in vivo
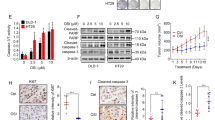
To analyze the in vitro effects of the oridonin on colon cancer cells, we treated several cell lines with different doses of oridonin for 24 h and assessed the effects on proliferation and survival. Significant cytotoxic activity was observed in all six CRC cell lines (HCT-15, COLO205, HCT116, RKO, SW480, and SW620) in a dose-dependent manner, with IC50 values ranging between 10 and 32 μM at 24 h (Figures 1 a and b). Then, SW480 and RKO cells were analyzed by flow cytometry following Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) dual labeling after the 24 h oridonin treatment. Results indicated that cell death rate is upregulated with oridonin concentration in both SW480 and RKO cells, but no typical phenotype of apoptosis was observed in p53-mutated CRC cell SW480, similar results were also detected in another p53-mutated CRC cell HCT-15 (Supplementary Figure 1), whereas in p53 wild-type (WT) CRC cell RKO showed classical apoptosis (Figure 1c). To confirm these results, we performed Western blots to measure the protein levels of caspase 3 and PARP, the most important molecular biomarkers of apoptosis. The results indicated that both caspase 3 and PARP were not significantly activated, and oridonin did not significantly induce apoptosis in SW480 cells (Figure 1d). But clearly caspase 3 activation and PARP cleavage were detected in RKO cells under oridonin treatment (Supplementary Figure 2). Animal xenograft studies indicated that tumor growth significantly delayed in nude mice treated with oridonin, and tumor weight was also significantly decreased compared with that of the solvent control group (Figures 1e and f, P<0.05), whereas the body weight changes showed no apparent differences between the treatment and control groups (data not shown). Collectively, these results indicated that oridonin exhibits potent anticancer effects on CRC cells both in vitro and in vivo, and it induced cell death through an atypical form of apoptosis in p53-mutated CRC cells.
Oridonin inhibits CRC cell proliferation and induces cell death both in vitro and in vivo. (a) Cell viability detected by CCK-8 assays in six CRC cell lines treated with different doses of oridonin for 24 h. (b) IC50 values of the oridonin in six CRC cells. (c) Contour diagrams of flow cytometry analysis of SW480 cells after the 24 h treatment; (d), Western blot analysis showing caspases were not activated following oridonin treatment. (e and f) Oridonin decreased tumor growth in a SW480 xenograft model as determined by assessing both tumor volume and tumor weight
Cellular energy homeostasis is altered by oridonin
To investigate the anticancer mechanisms of oridonin, we performed high-throughput sequencing to analyze miRNA expression, as miRNA expression profiling can be used to assess the responses to cancer therapies.21 The expression of 14 miRNAs reported to regulate key enzymes of the Warburg effect22 was significantly changed upon oridonin treatment in the SW480 cell line (Figure 2a). The results showed that MCT1-related miRNAs (miR-124-3p, miR-29b-1-5p, and miR-29a-5p) had the most significant expression changes, with more than two-fold increases, and the GLUT1 gene had the greatest number of related miRNAs (miR-130b-5p, miR-532-5p, miR-138-5p, miR-150-5p, miR-19b-3p, and miR-19a-3p) that were altered following oridonin treatment. Other miRNAs, including miR-378a-3p/5p targeting PGC-1, miR-200a/b/c-3p and miR-200b-5p targeting GPI, miR-125a-5p targeting HK2, and miR-320a targeting PKFM, also showed expression changes after oridonin treatment. Then, the expression of several representative miRNAs was confirmed by qRT-PCR (Figure 2b). From these results we can hypothesis that oridonin may affect cancer cell metabolism through those proteins involved in the Warburg effect.
Oridonin affects intracellular energy homeostasis. (a) A histogram showing differential expression of miRNAs related to glucose metabolism following oridonin treatment. (b) Verification of miRNAs by qPCR analysis. (c) Glucose uptake was inhibited by oridonin in SW480 cells. (d and e) Oridonin inhibited intracellular lactate export after a 24 h oridonin treatment. (f) Intracellular ATP levels were upregulated after the 24 h oridonin treatment. (g) ATP analysis during a time course experiment. ATP was significantly upregulated after a 12 h oridonin treatment. (**P<0.05)
To confirm this hypothesis, we investigated the intracellular energy status to assess the relationships between oridonin and cancer cells. First we evaluated the differences in glucose uptake, cellular lactate rate, and ATP level upon oridonin treatment in CRC cells. Glucose consumption of the SW480 cells significantly decreased after oridonin stimulation (Figure 2c). Furthermore, intracellular lactate concentration was upregulated, but extracellular lactate decreased in SW480 cells after 24 h of oridonin treatment suggesting lactate exportation is inhibited (Figures 2d and e). Intracellular ATP levels significantly increased by more than two-fold after the 24 h oridonin treatment in a dose-dependent manner (Figure 2f). To further elucidate how the ATP level increases, ATP levels were examined under 20 μM oridonin treatment in different times (0 h, 2 h, 4 h, 6 h, 8 h, 12 h, 16 h, and 24 h). The ATP level was not significantly different at these time points, except the 12 h time point, which showed a substantial ATP increase of more than three-fold (Figure 2g). Overall, these results suggested that oridonin could reduce glucose uptake, inhibit lactate export, and influence the cellular ATP supply to regulating cellular energy homeostasis.
Oridonin regulates cellular energy flow through GLUT1 and MCT1
The above findings suggesting us that some of the functional proteins and enzymes that regulate these processes in glycolytic metabolism were affected by oridonin. To verify this hypothesis, we first analyzed various proteins, including the GLUT1 and MCT1, which have been reported to be major regulators of the glucose supply and lactate export in colorectal carcinoma.23, 24 The results showed that the GLUT1 and MCT1 protein levels were downregulated following oridonin treatment for 24 h in a dose-dependent manner (Figure 3a). However, the protein level of LDHA, which is a crucial enzyme in controlling glycolysis, did not show similar results to those of GLUT1 and MCT1 (Figure 3a). Then, mRNA expression of these proteins was analyzed by qRT-PCR, and the results indicated that oridonin downregulated the mRNA expression of GLUT1, MCT1, and LDHA (Figure 3c). In addition, SW480 xenograft tumors treated with oridonin showed substantially reduced levels of GLUT1 and MCT1 compared with those of the control group, which confirmed the in vitro results (Figure 3b). Taken together, the data showed that oridonin affected glucose metabolism through the regulation of an energy supplier (GLUT1) and waste exporter (MCT1) but not the rate-limiting enzyme LDHA, which controls glycolytic flux.
Oridonin decreased the expression of glycolysis-related proteins. (a) Oridonin did not affect the expression of LDHA but caused a dose-dependent decrease of GLUT1 and MCT1 following 24 h oridonin treatment. (b) Immunohistochemistry analysis of GLUT1 and MCT1 in SW480 xenograft tumors. The sections were developed by diaminobenzidine and counterstained with hematoxylin. (c) Oridonin downregulated mRNA expression of glut1, mct1, and ldha in SW480 cells
Oridonin induces autophagy through deactivation of AMPK and downregulation of GLUT1 in SW480 cells
The above observations indicated that the intracellular ATP level was upregulated after a 24 h oridonin treatment, and the time course showed that ATP was upregulated at only a few time points during oridonin treatment. This result was unexpected as we had demonstrated that oridonin could downregulate the GLUT1 and MCT1 expression to reduce glucose consumption and decrease the lactate production of the cell, which indicating intracellular ATP level should decrease after oridonin treatment. There are four ATP sources in normal cells, including oxidative phosphorylation, glycolysis, glutaminolysis, and autophagy.25 The phenotype of the cancer cells indicated that oxidative phosphorylation and glutaminolysis cannot be the sources of the high intracellular ATP levels. Our finding of glucose uptake inhibition also suggested that glycolysis was not responsible for the ATP upregulation. Thus, we excluded three of the ATP sources and hypothesized that oridonin treatment induced cancer cell autophagy, resulting in intracellular ATP upregulation in SW480 cells.
To confirm our hypothesis, we first analyzed the expression of LC3, the cleaved and lipidated form (LC3-II) of which is widely regarded as an autophagosome marker. As shown in Figure 4a, the expression of LC3-II was upregulated following oridonin treatment in a dose-dependent manner. Then, transmission electron microscopy analysis was employed for a more detailed examination of the autophagy. Clear morphological characteristics of autophagosomes were observed in oridonin-treated SW480 cells, but no such phenomena were observed in the control group (Figure 4b). Some other autophagy-related proteins including beclin-1 and Atg5 could also by upregulated by oridonin treatment in HCT-15 cells, whereas oridonin could not induce those proteins in HCT116 cells (Supplementary Figure 3). In nude mice, oridonin could also upregulate the LC3-II expression in SW480 cells, accordingly (Supplementary Figure 4). These results verified our hypothesis that oridonin induced autophagy in p53 mutant type (MT) cells.
Oridonin could induce autophagy in a GLUT1-related but AMPK activation-independent manner. (a) Oridonin activated the LC3-II expression in SW480 cells after a 24 h treatment. (b) The ultrastructure of SW480 cells was observed by electron microscopy after oridonin treatment. Clear autophagosomes were observed in the oridonin treatment group, whereas this was not found in the control group. (c) AMPK was deactivated by oridonin as there was a decrease in p-AMPK in SW480 cells. (d) Detection of these proteins in a time course experiment after oridonin treatment. Oridonin downregulated p-AMPK and GLUT1 in a time-dependent manner in SW480 cells. (e) The autophagy inhibitor 3-MA (5 mmol/l) could alleviate the cytotoxicity of oridonin in SW480 cells. (f) Blocking AMPK by siRNA downregulated GLUT1 and induced LC3-II expression
AMPK, an important energy regulator in the cell that controls the expression of GLUT1 and MCT1, and promotes autophagy were detected. p-AMPK levels were substantially decreased with oridonin treatment, but total AMPK protein was unchanged (Figure 4c). Then, a time course experiment was carried out to assess the effects over time, and the results showed that oridonin could rapidly and significantly deactivate p-AMPK in <2 h, GLUT1 expression decreased in <4 h, whereas LC3-II levels were upregulated (Figure 4d). The LC3-II level also upregulated when we blocked GLUT1 with a specific inhibitor, WZB117, whereas similar results were observed when we knockdown AMPK expression, accompanied by a reduction in the GLUT1 level (Figure 4e). To further confirm oridonin-induced cell death via autophagy, by using a autophagy inhibitor 3-MA to block this process, we analyzed cell viability to confirm the autophagy in oridonin-induced cell death. The results revealed that 3-MA could alleviate the viability inhibition by oridonin treatment (Figure 4f).
Collectively, these results suggest that oridonin induced autophagy through downregulation of AMPK-GLUT1 and caused cell death in colorectal cancer cells.
Discussion
Over the past decade, the mechanisms underlying the anti-tumor activities of oridonin have been widely studied to provide support for the use of oridonin in clinical cancer therapy.21, 26, 27, 28, 29 Most studies, however, have not elucidated the detailed functions, mechanisms, and signaling pathways of its anticancer effect on solid tumors. Therefore, we conducted the present study to clarify the possible relationships between CRC cells and oridonin, and to explore the potential mechanisms of the anti-tumor activities of oridonin.
The cytotoxic activity and apoptosis induction of oridonin have been reported in several CRC cell lines including HT-29, HCT116, SW480, and SW620.16, 28, 30, 31, 32 But a very interesting finding grabbed our attention that although clear anti-proliferative activity was observed both in vitro and vivo, no typical apoptosis phenomenon was observed in SW480 cells, whereas clear apoptosis was detected in RKO cells using flow cytometry with Annexin V-FITC/PI labeling (Figure 1c). Thus, we hypothesized that oridonin caused cell death via a distinct process that was different from normal apoptosis in SW480 cells. Apoptosis biomarker analysis CRC cells (Figure 1d and Supplementary Figure 2) supported our hypothesis. These results were unexpected as apoptosis is considered to be the primary mechanism of oridonin anticancer activity, similar to what was observed in RKO cells and some previous reports.21, 28 There are three major types of morphologically distinct cell death: apoptosis (type I cell death), autophagic cell death (type II), and necrosis (type III).33 Here we did not observe the loss of the plasma membrane, which is characteristic of necrotic cell death. Thus, we could exclude both type I and type III cell death. Then, autophagic cell death could be the key mechanism underlying oridonin-induced cell death in SW480 and HCT-15 cells. This hypothesis was also supported by the results of our autophagy analysis. The results obtained in RKO and HCT116 cells suggested that oridonin promotes anti-tumor activity in both apoptotic and non-apoptotic manners. Previous studies have found that WT P53 protein was expressed in RKO and HCT116 cells, whereas SW480 and HCT-15 cells only expressed the MT P53 proteins. Studies indicated that CRC cells with MT P53 were more chemo-resistant than those with WT p53 due to their poor ability to activate apoptosis and initiation of apoptosis is the most important functions of P53.34, 35 Thus, we hypothesized that oridonin preferentially induced cell death through apoptosis in WT P53 cancer cells, whereas in those MT P53 cancer cells oridonin triggered another mechanism (e.g., autophagy) to compensate for the low apoptotic activity. Our study in those CRC cells supported our hypothesis, but further research is still needed to conduct the analysis in a more comprehensive way.
Accumulating evidence suggested that miRNA expression profiling could be used to elucidate various processes, such as apoptosis, autophagy,36 immunity,37 and cellular metabolism,22, 38, 39 in response to therapy in human cancers. Combining our data from high-throughput sequencing with the data reported in previous studies suggests that oridonin may influence the Warburg effect and affect its role through a metabolism-regulated pathway. A key finding in our study is that oridonin could decrease the glucose uptake and induce intracellular lactate accumulation (Figures 2c and d). Thus, we concluded that oridonin blocked the metabolic flux and inhibited anaerobic glycolysis, affecting the Warburg effect of the cancer cells. The Warburg effect describes the high flux of glucose through glycolysis, which converts hexoses to trioses, and results in pyruvate which converted to lactate.40 This whole reaction proceeds in the cytoplasm to produce a net 2 ATP from each glucose molecule, which is far less than that of the Krebs cycle, which generates 36 or 38 ATP. Due to the low efficiency of energy generation, cancer cells rely on the high rate of glucose consumption and lactate exportation to maintain the cellular energy supply and homeostasis. Targeting glucose metabolism of cancer cell has been shown to be a successful therapeutic strategy.41 This strategy not only altered cancer initiation and progression but also improved existing approaches, including diagnostic, drug resistance, or side effect reduction strategies, and is now a popular topic in cancer research.5, 41, 42 This study is the first to show that oridonin could target glucose metabolism, and it increases our knowledge of the effects of oridonin.
While it is clear that oridonin could affect intracellular energy flux, the underlying mechanism of how oridonin targets glucose metabolism is still under investigation. Various strategies aimed at regulating glucose metabolism are being researched and generally focus on three steps: 1. targeting glucose transport (e.g., Gluts) to control the cellular energy supply; 2. targeting rate-limiting enzymes (e.g., HK2, PKM2) to influence the glycolytic pathway; and 3. targeting lactate generation and export (e.g., MCTs, LDHA) to block the final stage of glycolysis. Our data showed that oridonin significantly downregulated GLUT1 and MCT1 expression in CRC cells. GLUT1 is one of the most important regulators of glucose import, shows variable levels in many tissues and is believed to be responsible for basal glucose uptake,24 whereas MCT1 plays a major role in transport of lactate, pyruvate, and other short-chain monocarboxylates across the membrane in a freely reversible manner.43 Thus, our results indicated that oridonin can block both energy supply and lactate export through the regulation of GLUT1 and MCT1 in CRC cells. Both GLUT1 and MCT1 expression are essentially ubiquitous across all mammalian tissues at low levels, whereas in most cancer cells, they are highly expressed.2, 44 The overexpression of GLUT1 is correlated to hypoxia, poor prognosis, and drug resistance in CRC.24, 45, 46, 47 Other reports found a similar relationship between MCT1 and various CRC characteristics including drug resistance, poor prognosis, and hypoxia.48, 49 However, the most important biological functions of these two proteins are to supply carbohydrates and export the waste lactate to maintain cellular energy homeostasis.50, 51, 52
The ATP increase observed after oridonin treatment (Figure 2f) was unexpected, as we had demonstrated that oridonin could block glucose uptake and lactate export. We predicted that the ATP level would decrease along with the reduced glucose levels. There are four major ATP supply methods – oxidative phosphorylation, glutaminolysis, autophagy, and glycolysis.25, 53 Combining our results shown in Figure 2c and the essential characteristics of cancer cells, we concluded that because cancer cells only maintain low levels of oxidative phosphorylation, glycolysis was blocked by the decrease in glucose, and glutaminolysis can only produce low levels of ATP, and autophagy was the likely mechanism of ATP upregulation caused by oridonin treatment. The results of autophagy-related biomarker analysis and transmission electron microscopy analysis confirmed the hypothesis (Figure 4; Supplementary Figure 3) that autophagy is induced by oridonin in p53-mutated CRC cells. In addition, this was supported by the results of flow cytometry analysis (Figure 1c; Supplementary Figure 1). Although induction of autophagy by oridonin has been reported for years in several cell lines, this is the first time a similar phenomenon has been reported in CRC cell lines.
Glucose metabolic disorders are intimately related to cellular autophagy, glucose deprivation, and decreased cellular levels of ATP, leading to activation of autophagy.54 To determine exactly how oridonin triggers autophagy, we analyzed ATP levels and related proteins in a time course experiment. The ATP levels only increased (Figure 2g) for a few hours after the detection of LC3-II activation (Figure 4d), which is a marker of autophagy. Thus, we concluded that autophagy is responsible for the ATP upregulation. Furthermore, a reduction in GLUT1 accompanied the activation of LC3-II (Figure 4d), suggesting that GLUT1 was correlated with the activation of autophagy. This is consistent with a recent report indicating that downregulation of GLUT1 was an important trigger of autophagy.55 Interestingly, it has been reported that activation of AMPK could promote autophagy.56 However, in our study, the rapid deactivation of AMPK indicated that oridonin induced autophagy independent of AMPK activation. As AMPK is upstream of GLUT1 and MCT1,51, 57 the depletion of p-AMPK contributed to the oridonin-induced downregulation of GLUT1 and MCT1. In addition, a recent report indicated that AMPK inhibition could also be a trigger to activate autophagy,58 which supports our conclusion that oridonin induces autophagy through AMPK deactivation-mediated GLUT1 downregulation, as shown in Figure 4f. Autophagy is an important process in response to cellular stress and generally acts as a pro-survival mechanism for the maintenance of cellular homeostasis following environmental stimuli, such as endoplasmic reticulum stress or hypoxic stress.59, 60 Inhibition of autophagy reduced the oridonin-induced anti-proliferative activity (Figure 4e), indicating that the autophagy induced by oridonin did not typically promote cell survival but instead a mechanism called autophagy-mediated cell death (which could involve a standard mechanism of cell death, such as apoptosis, but is triggered by autophagy).61
In summary, the results of these studies support the hypothesis that oridonin altered intracellular energy homeostasis and induced autophagy. We demonstrated the potent anticancer activity of oridonin in vitro and in vivo in CRC cells and determined that the underlying mechanism of oridonin involves induction of metabolic disorders and autophagy. Oridonin significantly downregulated GLUT1 and MCT1, and altered energy homeostasis in cancer cells. Meanwhile, deactivation of AMPK induced GLUT1 downregulation, and metabolic disorders were related to oridonin-mediated autophagy. Moreover, our data suggested that autophagy is associated with the anti-tumor activity induced by oridonin in CRC. These mechanisms are summarized in Figure 5.
Materials and methods
Cell lines and cell cultures
All six CRC cell lines (HCT-15, COLO205, HCT116, RKO, SW480, and SW620) were obtained from the cell bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI 1640 or DMEM containing 10% fetal bovine serum (HyClone, SV30160, Logan, UT, USA), 100 U/ml penicillin, and 100 mg/ml streptomycin in a humidified atmosphere containing 5% CO2 at 37 °C.
Cell viability and apoptosis assays
The cytotoxicity of oridonin in the CRC cells was assessed using a Cell Counting Kit-8 (Dojindo, Laboratories, Kumamoto, Japan). All CRC cells were seeded in 96-well microplates with appropriate numbers overnight and were then incubated at 37 °C with oridonin for 24 h. The optical density (OD) was measured by an ELISA reader at 450 and 630 nm. The percentage of cytotoxicity was calculated by the following equation: Cytotoxicity (%)=(OD control group × OD treatment group)/OD control group × 100%.
A FACS Calibur flow cytometer (BD Biosciences, San Jose, CA, USA) was used to analyze apoptosis with FITC-Annexin V/PI double staining. The data were analyzed by FlowJo software (TreeStar, Ashland, OR, USA).
Measurement of intracellular ATP and l-lactate levels
Intracellular ATP was measured using CellTiter-Glo Reagent (Promega, Madison, WI, USA). Briefly, cells were harvested, quantified, and then lysed with lysis buffer. Fifty microliters from each diluted sample was mixed with an equal volume of CellTiter-Glo Reagent, and the sample was incubated at room temperature for 10 min and analyzed using a GloMax luminometer (GloMax 96 Microplate Luminometer; Promega).
After oridonin treatment, the medium was removed from the cells, and lactate levels in the extracellular medium were measured using a l-lactate Colorimetric Assay Kit (Abcam, Cambridge, UK). Then, the intracellular lactate levels were measured in the cell lysates. Data were normalized to the final cell counts.
RNA extraction and real-time quantitative PCR
Total RNA was extracted using the TRIzol reagent (Invitrogen, CA, USA). First-strand cDNA was synthesized with Prime Script RT Master Mix (TaKaRa, Dalian, China). Then, quantitative PCR was conducted using SYBR Green PCR Master mix (TaKaRa) on a Bio-Rad Real-Time PCR instrument (Bio-Rad, CA, USA). The sequences of the primers were as follows (in the 5′ to 3′ orientation): GLUT1 forward, 5′-CTT TGT GGC CTT CTT TGA AGT-3′; GLUT1 reverse, 5′-CCA CAC AGT TGC TCC ACA T-3′; MCT1 forward, 5′-GGT GTCATT GGA GGT CTT GGG-3′; MCT1 reverse, 5′-GGC CAA TGG TCGCTTCTT G-3′; GAPDH forward, 5′-TGA CCT GCC GTC TAG AAA AAC C-3′; GAPDH reverse, 5′-GCC AAA TTC GTT GTC ATA CCA GG-3′; LDHA forward, 5′-ATCTTG ACC TAC GTG GCT TGG A-3′; LDHA reverse, 5′-CCA TAC AGG CAC ACTGGA ATC TC-3′. The data were analyzed by StepOne software (Applied Biosystems, Waltham, MA, USA) with GAPDH as the constitutive marker.
Western blot
Cells were rinsed twice with cold PBS and homogenized with RIPA lysis buffer (Cell Signaling Technology, Beverly, MA, USA), and a protease inhibitor cocktail (Roche, Berlin, Germany) was added according to the manufacturer’s recommendation (40 μg per sample). Proteins were separated by 10% SDS-PAGE and electrophoretically transferred onto a polyvinylidene fluoride membrane (Millipore, Temecula, CA, USA). The membrane was washed with TBST, blocked with 5% skimmed milk, diluted in TBST for 1 h and then incubated with appropriate primary antibodies (anti-p-AMPK, GAPDH, caspase 3, PARP, LDHA, LC3, and β-actin obtained from Cell Signaling Technology, anti-GLUT1 and MCT1 obtained from Millipore, and all the antibodies were diluted 1:1000) overnight at 4 °C. The membranes were then washed for 5 min three times with TBST, and primary antibodies were detected with goat anti-rabbit IgG (CST) antibodies conjugated to horseradish peroxide. Bands were visualized with the ECL Western Blot Detection System (CST) and subjected to autoradiography using X-ray film.
Animal models
A SW480 xenograft-bearing mouse model was constructed using 6-week-old BALB/c nude mice (Sun Yat-sen University). Each mouse was subcutaneously injected with 6 × 106 SW480 cells. When the volume of the tumors reached 140 mm3, the mice were randomly assigned to two groups and treated intraperitoneally with oridonin (15 mg/kg) or vehicle (2% DMSO). All mice were killed after a 12-day treatment, and the tumor volumes (length × width2 × 0.5236) and body weights were determined. Then, the tumors were collected for immunohistochemistry analysis.
Transmission electron microscopy analysis
Cells were washed with PBS and fixed in 3% glutaraldehyde dissolved in 0.1 mol/l phosphate buffer for 30 min at room temperature. The cells were then postfixed for 1 h in 1% OsO4 (Sigma-Aldrich, St. Louis, MO, USA). After dehydration, the cells were embedded in Epon 812 (SPI Supplies, West Chester, PA, USA) and polymerized at 60 °C for 24 h. EM images were acquired from thin sections using a JEM1400 electron microscope (JEOL, Tokyo, Japan). Images were digitally acquired from a randomly selected pool under each condition.
Statistical analyses
The results are expressed as the mean±S.D. Comparisons were made to determine significant differences between individual groups by ANOVA. All tests were performed using SPSS 19 (SPSS Inc., Chicago, IL, USA), and P<0.05 was considered significant. All experiments were performed at least three times.
References
Koppenol WH, Bounds PL, Dang CV . Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer 2011; 11: 325–337.
Adekola K, Rosen ST, Shanmugam M . Glucose transporters in cancer metabolism. Curr Opin Oncol 2012; 24: 650–654.
Flaveny CA, Griffett K, El-Gendy Bel D, Kazantzis M, Sengupta M, Amelio AL et al. Broad anti-tumor activity of a small molecule that selectively targets the Warburg effect and lipogenesis. Cancer Cell 2015; 28: 42–56.
Watson J . Oxidants, antioxidants and the current incurability of metastatic cancers. Open Biol 2013; 3: 120144.
Zhao Y, Butler EB, Tan M . Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis 2013; 4: e532.
Cheong H, Lu C, Lindsten T, Thompson CB . Therapeutic targets in cancer cell metabolism and autophagy. Nat Biotechnol 2012; 30: 671–678.
Levine AJ, Puzio-Kuter AM . The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 2010; 330: 1340–1344.
Liu Q, Chen L, Hu L, Guo Y, Shen X . Small molecules from natural sources, targeting signaling pathways in diabetes. Biochim Biophys Acta 2010; 1799: 854–865.
Flati V, Pasini E, D'Antona G, Speca S, Toniato E, Martinotti S . Intracellular mechanisms of metabolism regulation: the role of signaling via the mammalian target of rapamycin pathway and other routes. Am J Cardiol 2008; 101: 16E–21E.
Lee YS, Kim WS, Kim KH, Yoon MJ, Cho HJ, Shen Y et al. Berberine, a natural plant product, activates AMP-activated protein kinase with beneficial metabolic effects in diabetic and insulin-resistant states. Diabetes 2006; 55: 2256–2264.
Liu Y, Cao Y, Zhang W, Bergmeier S, Qian Y, Akbar H et al. A small-molecule inhibitor of glucose transporter 1 downregulates glycolysis, induces cell-cycle arrest, and inhibits cancer cell growth in vitro and in vivo. Mol Cancer Ther 2012; 11: 1672–1682.
Ikezoe T, Chen SS, Tong XJ, Heber D, Taguchi H, Koeffler HP . Oridonin induces growth inhibition and apoptosis of a variety of human cancer cells. Int J Oncol 2003; 23: 1187–1193.
Fujita E, Nagao Y, Node M, Kaneko K, Nakazawa S, Kuroda H . Antitumor activity of the Isodon diterpenoids: structural requirements for the activity. Experientia 1976; 32: 203–206.
Li X, Li X, Wang J, Ye Z, Li JC . Oridonin up-regulates expression of P21 and induces autophagy and apoptosis in human prostate cancer cells. Int J Biol Sci 2012; 8: 901–912.
Zhang YH, Wu YL, Tashiro S, Onodera S, Ikejima T . Reactive oxygen species contribute to oridonin-induced apoptosis and autophagy in human cervical carcinoma HeLa cells. Acta Pharmacol Sin 2011; 32: 1266–1275.
Zhu Y, Xie L, Chen G, Wang H, Zhang R . Effects of oridonin on proliferation of HT29 human colon carcinoma cell lines both in vitro and in vivo in mice. Pharmazie 2007; 62: 439–444.
Li CY, Wang EQ, Cheng Y, Bao JK . Oridonin: an active diterpenoid targeting cell cycle arrest, apoptotic and autophagic pathways for cancer therapeutics. Int J Biochem Cell Biol 2011; 43: 701–704.
Cheng Y, Qiu F, Ikejima T . Molecular mechanisms of oridonin-induced apoptosis and autophagy in murine fibrosarcoma L929 cells. Autophagy 2009; 5: 430–431.
Zang L, He H, Xu Q, Yu Y, Zheng N, Liu W et al. Reactive oxygen species H(2)O(2) and OH, but not O(2)(-) promote oridonin-induced phagocytosis of apoptotic cells by human histocytic lymphoma U937 cells. Int Immunopharmacol 2013; 15: 414–423.
Zhou GB, Kang H, Wang L, Gao L, Liu P, Xie J et al. Oridonin, a diterpenoid extracted from medicinal herbs, targets AML1-ETO fusion protein and shows potent antitumor activity with low adverse effects on t(8;21) leukemia in vitro and in vivo. Blood 2007; 109: 3441–3450.
Huang HL, Weng HY, Wang LQ, Yu CH, Huang QJ, Zhao PP et al. Triggering Fbw7-mediated proteasomal degradation of c-Myc by oridonin induces cell growth inhibition and apoptosis. Mol Cancer Ther 2012; 11: 1155–1165.
Jin LH, Wei C . Role of microRNAs in the Warburg effect and mitochondrial metabolism in cancer. Asian Pac J Cancer Prev 2014; 15: 7015–7019.
Lambert DW, Wood IS, Ellis A, Shirazi-Beechey SP . Molecular changes in the expression of human colonic nutrient transporters during the transition from normality to malignancy. Br J Cancer 2002; 86: 1262–1269.
Chung FY, Huang MY, Yeh CS, Chang HJ, Cheng TL, Yen LC et al. GLUT1 gene is a potential hypoxic marker in colorectal cancer patients. BMC Cancer 2009; 9: 241.
Oronsky BT, Oronsky N, Fanger GR, Parker CW, Caroen SZ, Lybeck M et al. Follow the ATP: tumor energy production: a perspective. Anticancer Agents Med Chem 2014; 14: 1187–1198.
Zhen T, Wu CF, Liu P, Wu HY, Zhou GB, Lu Y et al. Targeting of AML1-ETO in t(8;21) leukemia by oridonin generates a tumor suppressor-like protein. Sci Transl Med 2012; 4: 127ra138.
Ikezoe T, Yang Y, Bandobashi K, Saito T, Takemoto S, Machida H et al. Oridonin, a diterpenoid purified from Rabdosia rubescens, inhibits the proliferation of cells from lymphoid malignancies in association with blockade of the NF-kappa B signal pathways. Mol Cancer Ther 2005; 4: 578–586.
Kwan HY, Yang Z, Fong WF, Hu YM, Yu ZL, Hsiao WL . The anticancer effect of oridonin is mediated by fatty acid synthase suppression in human colorectal cancer cells. J Gastroenterol 2013; 48: 182–192.
Gao FH, Hu XH, Li W, Liu H, Zhang YJ, Guo ZY et al. Oridonin induces apoptosis and senescence in colorectal cancer cells by increasing histone hyperacetylation and regulation of p16, p21, p27 and c-myc. BMC Cancer 2010; 10: 610.
Yang J, Jiang H, Wang C, Yang B, Zhao L, Hu D et al. Oridonin triggers apoptosis in colorectal carcinoma cells and suppression of microRNA-32 expression augments oridonin-mediated apoptotic effects. Biomed Pharmacother 2015; 72: 125–134.
Jin H, Tan X, Liu X, Ding Y . Downregulation of AP-1 gene expression is an initial event in the oridonin-mediated inhibition of colorectal cancer: studies in vitro and in vivo. J Gastroenterol Hepatol 2011; 26: 706–715.
Ji Z, Tang Q, Zhang J, Yang Y, Liu Y, Pan Y . Oridonin-induced apoptosis in SW620 human colorectal adenocarcinoma cells. Oncol Lett 2011; 2: 1303–1307.
Green DR, Llambi F . Cell death signaling. Cold Spring Harb Perspect Biol 2015; 7: a006080.
Chen J . The cell-cycle arrest and apoptotic functions of p53 in tumor initiation and progression. Cold Spring Harb Perspect Med 2016; 6: a026104.
Iacopetta B . TP53 mutation in colorectal cancer. Hum Mutat 2003; 21: 271–276.
Xu J, Wang Y, Tan X, Jing H . MicroRNAs in autophagy and their emerging roles in crosstalk with apoptosis. Autophagy 2012; 8: 873–882.
Smyth LA, Boardman DA, Tung SL, Lechler R, Lombardi G . MicroRNAs affect dendritic cell function and phenotype. Immunology 2015; 144: 197–205.
Chen B, Li H, Zeng X, Yang P, Liu X, Zhao X et al. Roles of microRNA on cancer cell metabolism. J Trans Med 2012; 10: 228.
Eichner LJ, Perry MC, Dufour CR, Bertos N, Park M, St-Pierre J et al. miR-378(*) mediates metabolic shift in breast cancer cells via the PGC-1beta/ERRgamma transcriptional pathway. Cell Metab 2010; 12: 352–361.
Dang CV . Rethinking the Warburg effect with Myc micromanaging glutamine metabolism. Cancer Res 2010; 70: 859–862.
Vander Heiden MG . Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov 2011; 10: 671–684.
Elf SE, Chen J . Targeting glucose metabolism in patients with cancer. Cancer 2014; 120: 774–780.
Halestrap AP, Price NT . The proton-linked monocarboxylate transporter (MCT) family: structure, function and regulation. Biochem J 1999; 343 (Pt 2): 281–299.
Pullen TJ, Sylow L, Sun G, Halestrap AP, Richter EA, Rutter GA . Overexpression of monocarboxylate transporter-1 (SLC16A1) in mouse pancreatic beta-cells leads to relative hyperinsulinism during exercise. Diabetes 2012; 61: 1719–1725.
Sakashita M, Aoyama N, Minami R, Maekawa S, Kuroda K, Shirasaka D et al. Glut1 expression in T1 and T2 stage colorectal carcinomas: its relationship to clinicopathological features. Eur J Cancer 2001; 37: 204–209.
Liu W, Fang Y, Wang XT, Liu J, Dan X, Sun LL . Overcoming 5-Fu resistance of colon cells through inhibition of Glut1 by the specific inhibitor WZB117. Asian Pac J Cancer Prev 2014; 15: 7037–7041.
Shang R, Wang J, Sun W, Dai B, Ruan B, Zhang Z et al. RRAD inhibits aerobic glycolysis, invasion, and migration and is associated with poor prognosis in hepatocellular carcinoma. Tumour Biol 2015; 37: 5097–5105.
Verstraete M, Debucquoy A, Devos E, Sagaert X, Penninckx F, Begg A et al. Investigation of possible endogenous hypoxia markers in colorectal cancer. Int J Radiat Biol 2013; 89: 9–15.
Amorim R, Pinheiro C, Miranda-Goncalves V, Pereira H, Moyer MP, Preto A et al. Monocarboxylate transport inhibition potentiates the cytotoxic effect of 5-fluorouracil in colorectal cancer cells. Cancer Lett 2015; 365: 68–78.
Cuff MA, Lambert DW, Shirazi-Beechey SP . Substrate-induced regulation of the human colonic monocarboxylate transporter, MCT1. J Physiol 2002; 539 (Pt 2): 361–371.
Wu N, Zheng B, Shaywitz A, Dagon Y, Tower C, Bellinger G et al. AMPK-dependent degradation of TXNIP upon energy stress leads to enhanced glucose uptake via GLUT1. Mol Cell 2013; 49: 1167–1175.
Granja S, Morais-Santos F, Miranda-Goncalves V, Viana-Ferreira M, Nogueira R, Nogueira-Silva C et al. The monocarboxylate transporter inhibitor alpha-cyano-4-hydroxycinnamic acid disrupts rat lung branching. Cell Physiol Biochem 2013; 32: 1845–1856.
Wang Y, Martins I, Ma Y, Kepp O, Galluzzi L, Kroemer G et al. Release from dying cells via lysosomal exocytosis. Autophagy 2013; 9: 1624–1625.
Kim KH, Lee MS . Autophagy—a key player in cellular and body metabolism. Nat Rev Endocrinol 2014; 10: 322–337.
Buller CL, Heilig CW, Brosius FC 3rd . GLUT1 enhances mTOR activity independently of TSC2 and AMPK. Am J Physiol Renal Physiol 2011; 301: F588–F596.
Liu X, Niu Y, Yuan H, Huang J, Fu L . AMPK binds to sestrins and mediates the effect of exercise to increase insulin-sensitivity through autophagy. Metabolism 2015; 64: 658–665.
Takimoto M, Takeyama M, Hamada T . Possible involvement of AMPK in acute exercise-induced expression of monocarboxylate transporters MCT1 and MCT4 mRNA in fast-twitch skeletal muscle. Metabolism 2013; 62: 1633–1640.
Liu X, Chhipa RR, Nakano I, Dasgupta B . The AMPK inhibitor compound C is a potent AMPK-independent antiglioma agent. Mol Cancer Ther 2014; 13: 596–605.
Fulda S, Kogel D . Cell death by autophagy: emerging molecular mechanisms and implications for cancer therapy. Oncogene 2015; 34: 5105–5113.
Kim JH, Kim HY, Lee YK, Yoon YS, Xu WG, Yoon JK et al. Involvement of mitophagy in oncogenic K-Ras-induced transformation: overcoming a cellular energy deficit from glucose deficiency. Autophagy 2011; 7: 1187–1198.
Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8: 445–544.
Acknowledgements
We would like to thank Qiao-Juan Huang, Yi-Ling Chen, and Xiao Hong Chen for their technical assistance. This work was supported by the National Natural Science Foundation of China (31230042, 31471223) and the project of Science and Technology of Guangzhou (201504010022).
Author contributions
Zhuo Yao and Fuhua Xie contributed equally to this work.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by GM Fimia
Supplementary Information accompanies this paper on Cell Death and Disease website
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Yao, Z., Xie, F., Li, M. et al. Oridonin induces autophagy via inhibition of glucose metabolism in p53-mutated colorectal cancer cells. Cell Death Dis 8, e2633 (2017). https://doi.org/10.1038/cddis.2017.35
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2017.35
This article is cited by
-
ThermomiR-377-3p-induced suppression of Cirbp expression is required for effective elimination of cancer cells and cancer stem-like cells by hyperthermia
Journal of Experimental & Clinical Cancer Research (2024)
-
Biophysical and biochemical properties of PHGDH revealed by studies on PHGDH inhibitors
Cellular and Molecular Life Sciences (2022)
-
Oridonin enhances antitumor effects of doxorubicin in human osteosarcoma cells
Pharmacological Reports (2022)
-
Aloin induced apoptosis by enhancing autophagic flux through the PI3K/AKT axis in osteosarcoma
Chinese Medicine (2021)
-
Terpenoids’ anti-cancer effects: focus on autophagy
Apoptosis (2021)