Abstract
Hepatocellular carcinoma (HCC) is characterized by a poor prognosis and is one of the leading causes of cancer-related death worldwide. Simvastatin, an HMG-CoA reductase inhibitor, which decreases cholesterol synthesis by inhibiting mevalonate pathways and is widely used to treat cardiovascular diseases. Simvastatin exhibits anticancer effects against several malignancies. However, the molecular mechanisms underlying the anticancer effects of simvastatin on HCC are still not well understood. In this study, we demonstrated simvastatin-induced G0/G1 arrest by inducing p21 and p27 accumulation in HepG2 and Hep3B cells. Simvastatin also promoted AMP-activated protein kinase (AMPK) activation, which induced p21 upregulation by increasing its transcription. Consistent with this finding, we found genetic silencing of AMPK reduced p21 expression; however, AMPK silencing had no effect on p27 expression in HCC cells. Simvastatin decreased Skp2 expression at the transcriptional level, which resulted in p27 accumulation by preventing proteasomal degradation, an effect mediated by signal transducer and activator of transcription 3 (STAT3) inhibition. Constitutive STAT3 activation maintained high-level Skp2 expression and lower level p27 expression and significantly prevented G0/G1 arrest in simvastatin-treated HCC cells. Mevalonate decreased simvastatin-induced AMPK activation and rescued phospho-STAT3 and Skp2 expression in HCC cells, which resulted in the prevention of G0/G1 arrest through inhibition of p21 and p27 accumulation. Moreover, simvastatin significantly decreased tumor growth in HepG2 xenograft mice. Consistently, we found that simvastatin also increased p21 and p27 expression in tumor sections by reducing Skp2 expression and inducing AMPK activation and STAT3 suppression in the same tumor tissues. Taken together, these findings are demonstrative of the existence of a novel pathway in which simvastatin induces G0/G1 arrest by upregulating p21 and p27 by activating AMPK and inhibiting the STAT3–Skp2 axis, respectively. The results identify novel targets that explain the beneficial anticancer effects of simvastatin treatment on HCC in vitro and in vivo.
Similar content being viewed by others
Main
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the third leading cause of cancer-related mortality worldwide.1, 2 HCC is induced by multiple conditions, including hepatitis B virus (HBV) infection, hepatitis C virus (HCV) infection, alcoholic liver disease and metabolic syndrome.3 Despite advances in HCC diagnosis and treatment, most HCC patients still have a poor prognosis because of tumor progression or tumor recurrence.4 Therefore, it is essential to develop chemopreventive strategies to improve HCC patient outcomes. In a previous study of patients with HBV- and HCV-related HCC, we reported that antiviral therapy reduced HCC recurrence and mortality after liver resection or radiofrequency ablation.5, 6, 7 However, antiviral therapy only reduces the risk of HCC. It does not eliminate the risk of the disease. Metabolic syndrome seems to have important roles in HCC development in the post-antiviral therapy era. We found that the risk of HCC was significantly higher in diabetic patients than in the general population and that metformin decreased the risk of HCC by inhibiting hepatoma cell proliferation and inducing cell cycle arrest.8 Statins have been suggested to inhibit HCC progression and increase HCC survival in patients with hyperlipidemia.
Statins are 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase inhibitors, which catalyze the rate-limiting step in cholesterol biosynthesis and are widely used to treat patients with hypercholesterolemia.9 The chemopreventive effects of statins have been reported in several cancers, such as gastric, breast, colon, liver and prostate cancer.10, 11, 12 Their potential mechanisms of action involve the inhibition of tumor cell proliferation, the promotion of cell cycle arrest, the induction of apoptotic cell death, and the inhibition of cell migration, invasion and metastasis.9, 10, 11, 12, 13 In addition, statins attenuate the production of isoprenoid units, which are critical for the activation of Rho, Ras and Rab proteins.12, 13 Statins also affect cyclin-dependent kinase inhibitors12 and have been shown to inhibit PI3K/AKT signaling and induce derepression of PTEN expression to inhibit breast cancer cell growth.14, 15 AMP-activated protein kinase (AMPK), a cellular energy sensor that mediates metabolic homeostasis under environmental stress conditions, was recently reported to be activated by statins.16, 17, 18, 19, 20 Autophagy inhibition enhances the anticancer effects of statins in digestive malignancies.18, 21 AMPK activation has been reported to suppress cell proliferation in non-tumor and tumor cells by regulating cell cycle progression or inhibiting protein synthesis.9, 22, 23, 24 In addition, signal transducer and activator of transcription 3 (STAT3), an important signaling protein that contributes to HCC development and progression, may be inhibited by statins in several cellular systems.25, 26, 27 The findings of recent studies suggest that simvastatin may inhibit cancer cell growth by inducing apoptosis or cell cycle arrest at the G0/G1 phase, thereby decreasing the risk of HCC.9, 28 However, whether AMPK and STAT3 have roles in the anticancer effects of statins in HCC remains unclear.
In this study, we showed that simvastatin can induce cell cycle arrest and increase cyclin-dependent kinase inhibitor expression in HCC cells. We demonstrated that simvastatin-induced cell cycle arrest was regulated by AMPK activation and STAT3 inactivation to transcriptionally increase p21 expression and stabilize p27 protein expression by inhibiting Skp2 expression, respectively. Moreover, we established a HepG2 tumor-bearing xenograft animal model to examine the antitumor effects of simvastatin in vivo. Our results showed that simvastatin inhibited tumor growth and that the expression patterns of p21, p27, Skp2, AMPK and STAT3 were similar to those of the in vitro study. Overall, our findings provide evidence of the existence of a novel molecular mechanism by which simvastatin exerts its anticancer effects in HCC.
Results
Simvastatin induces p21 and p27 expression-dependent G0/G1 cell cycle arrest in HCC cell lines
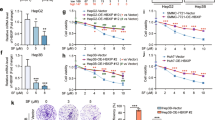
To determine whether simvastatin influences cell growth in hepatoma, we investigated the effect of simvastatin on cell viability in the HepG2 and Hep3B hepatoma cell lines. Simvastatin had significant dose- and time-dependent inhibitory effects on hepatoma cell growth in HepG2 and Hep3B cells, as demonstrated by CCK-8 assay (Figure 1a). To evaluate whether simvastatin induces cell death in hepatoma, we performed a viable cell count assay by Trypan blue staining in HepG2 and Hep3B cells. The results showed that the decrease in HepG2 and Hep3B cell viability elicited by 5–20 μg/ml simvastatin treatment (Figure 1a) did not correspond to the cell death rate of both cells subjected to the same dosage of simvastatin treatment (Figure 1b). Thus, simvastatin mainly inhibited HepG2 and Hep3B cell growth but did not cause cell death at doses <40 μg/ml. In addition, simvastatin treatment at a dose of 40 μg/ml induced higher apoptosis in HepG2 cells than in Hep3B cells (Supplementary Figure S1). The mechanism underlying statin-induced p53-dependent apoptosis has been clearly elucidated.29 Here, we focused on how simvastatin inhibits hepatoma cell growth. To investigate the mechanism underlying simvastatin-induced hepatoma cell growth inhibition, we analyzed the effect of simvastatin on cell cycle distribution using flow cytometry. We observed that after 48 h of incubation, simvastatin arrested cell cycle progression at the G0/G1 phase in HepG2 cells in a dose-dependent manner. The sizes of the sub-G1 populations of HepG2 cells were not significantly increased in the control and simvastatin-treated groups (Figure 1c). Simvastatin had a similar effect on cell cycle distribution in Hep3B cells (Figure 1d). As shown in Figure 1c, various doses of simvastatin arrested the cell cycle at the G0/G1 phase. The cell populations increased from 54.5% in control cells to 70.7% (20 μg/ml simvastatin) in HepG2 cells and from 42.74% in control cells to 59.2% (20 μg/ml simvastatin) in Hep3B cells. In addition, we observed that simvastatin treatment decreased cyclin D1 expression and increased p21 and p27 expression but had no significant modulatory effects on cyclin E1 expression in either HepG2 or Hep3B cells (Figure 1e). To determine whether simvastatin treatment-induced G0/G1 cell cycle arrest was dependent on p21 or p27 expression in HCC cells, we genetically silenced p21 and p27 in HepG2 cells and confirmed the decline of p21 and p27 expression by immunoblotting (Figure 1f). These p21 and p27 knockdown cells rescued the G0/G1 cell population under simvastatin treatment (Figure 1g). These results indicated that simvastatin treatment-induced G0/G1 cell cycle arrest that was dependent on p21 and p27 accumulation in HepG2 cells.
Simvastatin induces p21 and p27-dependent G0/G1 cell cycle arrest in HCC cell lines. Simvastatin suppressed cell growth in HCC cells. HepG2 and Hep3B cells were treated with various concentrations of simvastatin for 24 and 48 h. (a) Cell growth inhibition was measured by CCK-8 assay. (b) Cell death rates were determined by viable cell counting. These data are presented as percentages of vehicle-treated cells (*un-treated versus treated of HepG2 or Hep3B cells for 24 h or 48 h). (c and d) Simvastatin-induced HCC cell G0/G1 phase arrest. HepG2 and Hep3B cells were treated with simvastatin (0, 5, 10 or 20 μg/ml) for 48 h, and then cell cycle distributions were analyzed by PI staining and flow cytometry. (e) Simvastatin-induced G0/G1 phase-related protein expression in HCC cells. The cells were treated with simvastatin (0, 5, 10 or 20 μg/ml) for 24 h, and then the cell lysates were harvested for analysis of the expression of the cell cycle-related proteins p21, p27, cyclin D1, cyclin E1 and β-actin by immunoblotting. (f and g) Simvastatin-induced p21- and p27-dependent G0/G1 cell cycle arrest in HCC cells. HepG2 cells were transfected with p21, p27 or control siRNA for 24 h and then treated with 10 μg/ml simvastatin for 48 h. The cells were then harvested, and their DNA content and protein expression were analyzed by flow cytometry and immunoblotting using p21, p27 and β-actin antibodies. Data are expressed as the mean±S.E.M. of three independent experiments. Statistically significant differences between the un-treated and treated groups are indicated. *P<0.05, **P<0.01, ***P<0.001
Simvastatin induces p21 and p27 accumulation by increasing p21 transcription and preventing p27 degradation in HepG2 cells
To evaluate whether simvastatin increased p21 and p27 levels at the transcriptional, translational or degradation level, we first analyzed p21 and p27 mRNA levels by RT-PCR and real-time PCR after simvastatin treatment. We observed that p21 mRNA levels were upregulated and that p27 mRNA levels were not significantly affected by simvastatin stimulation (Figure 2a). We then examined whether simvastatin stabilized p21 and p27 protein expression. Compared with non-treated cells, simvastatin-treated cells exhibited a slower decrease in p27 expression than in p21 expression following treatment with the translation inhibitor cycloheximide (CHX) (Figure 2b). One of the mechanisms that controls p21 and p27 levels is proteasomal degradation.30, 31 We used MG132 to clarify whether simvastatin inhibits p21 and p27 protein degradation by blocking the ubiquitin proteasome pathway. We surmised that if simvastatin inhibits p21 and p27 proteasomal degradation, the protein levels of p21 and p27 will not change in the presence of MG132, irrespective of simvastatin administration. We thus also surmised that if simvastatin does not inhibit proteasomal degradation, the protein levels of p21 and p27 will increase upon simvastatin treatment in the presence of MG132, which would indicate that simvastatin either inhibits protein degradation by blocking another pathway, such as the lysosomal pathway, or induces protein synthesis at the transcriptional and/or translational level. Based on the results of this experiment, we determined that p21 levels increased significantly in the presence of MG132 following simvastatin treatment, but p27 levels were not affected by simvastatin treatment (Figure 2c). Taken together, our results suggested that simvastatin-induced p21 protein expression mainly at the transcription level and upregulated p27 protein expression predominantly by preventing protein degradation in HepG2 cells.
Simvastatin-induced p21 and p27 protein upregulation was associated with transcriptional activation and protein degradation inhibition, respectively. (a) Simvastatin modulated p21 mRNA expression. HepG2 cells were treated with simvastatin (0, 5, 10 or 20 μg/ml) for 24 h, and p21, p27 and GAPDH mRNA expression levels were detected by RT-PCR and real-time PCR. (b) The effects of p21 and p27 protein stability in simvastatin-treated HCC cells. HepG2 cells were treated with 10 μg/ml CHX alone for 1, 2, 4, 6 or 12 h or 10 μg/ml simvastatin for 12 h. After 12 h, simvastatin-treated cells were co-treated with 10 μg/ml CHX for 1, 2, 4, 6 or 12 h. The cell lysates were harvested to detect p21, p27 and β-actin protein expression by immunoblotting. The intensity of each protein signal was determined by ImageJ software (downloaded from the NIH website (http://rsb.info.nih.gov/ij)). (c) Inhibition of proteasomal degradation promoted p21 accumulation, but not p27 accumulation, in simvastatin-treated HCC cells. HepG2 cells were treated with 20 μg/ml simvastatin with or without 10 μM MG132 for 24 h and then subjected to immunoblotting for the detection of p21, p27 and β-actin expression levels. Data are expressed as the mean±S.E.M. of three independent experiments. Statistically significant differences between the un-treated and treated groups are indicated. **P<0.01, ***P<0.001
Simvastatin-induced p21 transcriptional upregulation is AMPK dependent in HepG2 cells
Simvastatin has been reported to activate the AMPK pathway.18, 19 To investigate whether AMPK has a role in influencing G0/G1 phase arrest in HepG2 cells after simvastatin treatment, we detected AMPK expression levels by immunoblotting analysis. Similar to the above results pertaining to the expression patterns of p21 and p27, these results showed that simvastatin treatment increased the level of phosphorylated AMPK (Figure 3a). To further examine whether AMPK-mediated simvastatin-induced G0/G1 phase arrest, we genetically knocked down AMPK expression in HepG2 cells using small interfering RNA (siRNA). AMPK knockdown cells displayed a partially but significantly decreased G0/G1 cell population under simvastatin treatment (Figure 3b). We also found that AMPK knockdown reduced p21 expression, but not p27 expression, in simvastatin-treated HepG2 cells (Figure 3c). These results provided evidence that AMPK activation by simvastatin-induced p21 expression at the transcriptional level and that p21 expression may be involved in simvastatin-induced cell cycle G0/G1 arrest in hepatoma cells.
Simvastatin-induced AMPK activation and p21 upregulation partially induced G0/G1 arrest in HepG2 cells. (a) Simvastatin-induced AMPK activation was associated with p21 and p27 upregulation. HepG2 cells were treated with simvastatin (0, 5, 10 or 20 μg/ml) for 12 h, and then immunoblotting was used to detect p-AMPK, AMPK, p21, p27 and β-actin protein expression levels. (b and c) Genetic silencing of AMPK reduced G0/G1 phase arrest and p21 expression in simvastatin-treated HCC cells. HepG2 cells were transfected with AMPK or control siRNA for 24 h and then treated with 10 μg/ml simvastatin for 48 h. (b) Cells were collected for analysis of their DNA content by flow cytometry. (c) Cell lysates were harvested to detect protein expression levels by immunoblotting using p-AMPK, AMPK, p21, p27 and β-actin antibodies. The results were obtained from three independent experiments. Data are expressed as the mean±S.E.M. of three independent experiments. **P<0.01, ***P<0.001
Simvastatin transcriptionally inhibits Skp2 expression and promotes p27 accumulation in HepG2 cells
Recent studies have shown that Skp2 E3 ligase activity can promote p27 degradation to prevent p27-induced cell cycle arrest.30, 32, 33 Therefore, we hypothesized that simvastatin may inhibit Skp2 to promote p27 accumulation and G0/G1 cell cycle arrest in HCC cells. As shown in Figure 4a, simvastatin decreased Skp2 protein expression in a dose-dependent manner. To evaluate the mechanism underlying simvastatin-induced decreases in Skp2 expression in HepG2 cells, we first analyzed Skp2 mRNA expression after simvastatin treatment using RT-PCR and real-time PCR. Our data showed that Skp2 mRNA expression decreased during simvastatin treatment (Figure 4b). We also confirmed that simvastatin inhibited Skp2 transcription in HepG2 cells by Skp2 promoter-driven luciferase assay (Figure 4c). Consistent with these findings, we found that Skp2 overexpression significantly prevented simvastatin-induced G0/G1 cell cycle arrest and p27 accumulation (Figures 4d and e). In addition, compared with control HepG2 cells, Skp2-overexpressing HepG2 cells did not maintain the simvastatin-enhanced p27 protein stability after treatment with CHX (Supplementary Figure S2). These results indicated that simvastatin treatment downregulated Skp2 expression at the transcriptional level and then promoted p27 accumulation to induce G0/G1 cell cycle arrest.
Simvastatin-induced p27 upregulation was Skp2-dependent and promoted G0/G1 phase cell cycle arrest in HepG2 cells. (a) Simvastatin decreased Skp2 protein expression in HepG2 cells. HepG2 cells were treated with simvastatin (0, 5, 10 or 20 μg/ml) for 24 h, and then cell lysates were harvested to detect the protein expression levels of Skp2 and β-actin by immunoblotting. (b) Simvastatin inhibited Skp2 mRNA expression in HepG2 cells. After the same treatment, cells were collected for analysis of Skp2 and GAPDH mRNA expression levels by RT-PCR and real-time PCR. (c) Simvastatin reduced Skp2 promoter activity. HepG2 cells were transfected with a pGL4.18-Skp2 promoter plasmid for 24 h and were then treated with simvastatin (0 or 20 μg/ml) for 24 h. The cell lysates were harvested to assay luciferase activity using a dual-luciferase assay kit. Data were normalized to Renilla luciferase activity and expressed as fold inductions of the control. (d) Skp2 overexpression rescued cells from simvastatin-induced G0/G1 cell cycle arrest. Control and Skp2-overexpressing HepG2 cells were treated with simvastatin (0, 10 or 20 μg/ml) for 48 h, and then the cells were collected for DNA content assay by flow cytometry. (e) Simvastatin was unable to change Skp2 and p27 protein expression levels in Skp2-overexpressing HepG2 cells. Control and Skp2-overexpressing HepG2 cells were treated with simvastatin (0 or 20 μg/ml) for 24 h, and then the cell lysates were collected for protein expression detection by immunoblotting using Skp2, p27 and β-actin antibodies. The results were obtained from three independent experiments. Data are expressed as the mean±S.E.M. of three independent experiments. **P<0.01, ***P<0.001
Simvastatin inhibits the STAT3/Skp2 axis to induce G0/G1 cell cycle arrest in HepG2 cells
It has been reported that STAT3 inactivation induces Skp2 downregulation and p27 upregulation in cervical and gastric cancer.34, 35 We investigated the molecular mechanisms underlying this phenomenon to determine whether STAT3 interacts with the Skp2/p27 pathway in simvastatin-treated HepG2 cells. We found that simvastatin decreased phospho-STAT3 levels, as well as those of its upstream regulators, Jak1 and Jak2, in HepG2 cells, as shown in Figure 5a. Next, we attempted to evaluate whether constitutive STAT3 activation facilitated by the expression of a constitutively activate mutant of STAT3 (STAT3C) could reverse the above simvastatin-induced effects. We transfected HepG2 cells with the STAT3C expression vector and selected cells that stably expressed STAT3C. We observed that the G0/G1 cell population in STAT3C-transfected cells was lower than that in mock cells after simvastatin treatment (Figure 5b). As shown in Figure 5c, the mRNA expression levels of Skp2 increased significantly in STAT3C-transfected cells compared with those in mock cells, confirming the existence of a relationship between STAT3 and Skp2. To further investigate whether cell cycle-related molecule expression levels were affected in the mock and STAT3C-transfected groups after simvastatin treatment, we detected p-STAT3, STAT3, Skp2 and p27 expression levels by immunoblotting analysis. We found that STAT3C-transfected cells treated with simvastatin maintained higher Skp2 protein expression levels but displayed severely decreased p27 expression compared with mock cells (Figure 5d). Thus, our results indicated that simvastatin-induced p27 upregulation is Skp2 dependent and occurs through inhibition of the STAT3/Skp2 activation axis.
STAT3C mutants maintained Skp2 expression to prevent p27 accumulation and G0/G1 cell cycle arrest in simvastatin-treated HepG2 cells. (a) Simvastatin inhibited the Jak1/Jak2-STAT3 pathway in HCC cells. HepG2 cells were treated with simvastatin (0, 5, 10 or 20 μg/ml) for 24 h, and then the cell lysates were harvested for the detection of protein levels by immunoblotting using p-Jak1, Jak1, p-Jak2, Jak2, p-STAT3, STAT3 and β-actin antibodies. (b) STAT3C mutants prevented simvastatin-induced G0/G1 cell cycle arrest in HepG2 cells. HepG2 cells were stably transfected with control vectors or STAT3C mutants and were then treated with simvastatin (0, 10 or 20 μg/ml) for 48 h. Then, the cells were collected for DNA content assay by flow cytometry. (c) Constitutive STAT3 activation in HepG2 cells upregulated Skp2 expression at the transcriptional level. Control and constitutive STAT3 activity levels in HepG2 cells were analyzed by RT-PCR to determine HepG2 Skp2 mRNA expression levels. (d) HepG2 cells stably expressing STAT3C maintained higher Skp2 levels and lower p27 levels. Control and STAT3C-expressing HepG2 cells were treated with simvastatin (0 or 20 μg/ml). Twenty-four hours later, the cell lysates were collected to detect protein expression by immunoblotting using p-STAT3, STAT3, Skp2, p27 and β-actin antibodies. The results were obtained from three independent experiments. Data are expressed as the mean±S.E.M. of three independent experiments. ***P<0.001
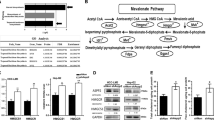
Mevalonate reverses the activation of AMPK and the inhibition of STAT3 facilitated by simvastatin treatment in HepG2 cells
Many studies have demonstrated that statins decrease cholesterol biosynthesis by inhibiting the endogenous mevalonate pathway.36, 37, 38 Here, restoration of mevalonate, the HMG-CoA reductase product, significantly reduced AMPK phosphorylation (Figure 6a) and maintained higher phospho-STAT3 and Skp2 levels, with decreased p21 and p27 accumulation, in simvastatin-treated HepG2 cells (Figure 6b). Moreover, we also observed that mevalonate treatment effectively prevented simvastatin-induced G0/G1 cell cycle arrest in HepG2 cells (Figure 6c). Taken together, our findings demonstrated that AMPK pathway activation and STAT3/Skp2 pathway inhibition in HepG2 cells are dependent on inhibition of the conversion of HMG-CoA reductase to mevalonate by simvastatin.
Exogenous mevalonate suppressed the inhibitory effects of simvastatin on cell growth by reversing the simvastatin-induced AMPK activation and STAT3 inhibition in HepG2 cells. (a) Mevalonate inhibited simvastatin-induced AMPK activation and p21 enhancement. HepG2 cells were pre-treated with or without mevalonate (3 mM) for 1 h and then treated with 20 μg/ml simvastatin for 12 h. The cells were collected to detect p-AMPK, AMPK, p21 and β-actin protein levels by immunoblotting. (b) Mevalonate blocked simvastatin-induced STAT3 inhibition and Skp2 depletion. HepG2 cells were pre-treated with or without mevalonate (3 mM) for 1 h and then treated with 20 μg/ml simvastatin for 24 h. The cell lysates were harvested to analyze protein levels by immunoblotting using p-STAT3, STAT3, Skp2, p27 and β-actin antibodies. (c) Mevalonate inhibited simvastatin-induced G0/G1 cell cycle arrest. HepG2 cells were pre-treated with or without mevalonate (3 mM) for 1 h, followed by treatment with 20 μg/ml simvastatin for 48 h. The cells were collected for analysis of DNA content by flow cytometry using PI staining. Results are shown as the mean±S.E.M. of three independent experiments. ***P<0.001
Simvastatin inhibits tumor growth in xenograft animal models
To examine the antitumor effect of simvastatin in vivo, we injected HepG2 cells subcutaneously into the flanks of BALB/c nude mice to establish a HepG2 tumor-bearing animal model. As shown in Figure 7a, we demonstrated that simvastatin significantly suppressed tumor growth (Figure 7b) and reduced tumor weight (Figure 7c) in nude mice. Consistent with these findings, we found that p21 and p27 expression levels were increased by simvastatin treatment in mice compared with saline treatment (Figure 7d). In addition, using immunohistochemistry (IHC), we also observed that simvastatin treatment increased AMPK phosphorylation, reduced STAT3 phosphorylation and decreased Skp2 expression in tumor tissues (Figure 7d). Taken together, these results indicated that simvastatin repressed tumor growth by increasing p21 and p27 expression through AMPK activation and STAT3/Skp2 axis inhibition in vivo.
Simvastatin inhibited HepG2 tumor growth in xenograft mice. HepG2 tumor-bearing BALB/c nude mice were divided into control and simvastatin treatment groups. The mice were treated with saline or 20 mg/kg body weight simvastatin twice a day by intraperitoneal (i.p.) injection. (a) Tumor tissues from the saline and simvastatin treatment groups were harvested at 14 days after injection. These tissues were isolated from each mouse after killing. (b) Tumor growth curves derived from nude mice in the saline- and simvastatin-treated groups. The tumor volumes of the nude mice were calculated twice a day for 2 weeks. (c) The tumor weights of the nude mice were measured after killing. Results are shown as the mean±S.E.M (n=12). Saline-treated mice compared with simvastatin-treated mice. ***P<0.001 (d) IHC analysis using antibodies against human p21, p27, Skp2, p-AMPK and p-STAT3 in the tumor tissues of the saline- and simvastatin-treated groups. All scale bars are 50 μm. (e) To summarize the in vitro and in vivo results of this study, we found that simvastatin promoted G0/G1 cell cycle arrest by increasing p21 and p27 expression via AMPK pathway activation and STAT3/Skp2 pathway suppression, respectively. These phenomena were dependent on inhibiting the production of mevalonate by simvastatin treatment in the HCC model
Discussion
In this study, we investigated the molecular mechanisms by which simvastatin induces cell growth arrest in HCC cells and found that simvastatin treatment results in suppression of the oncoproteins STAT3 and Skp2 and activation of the energy sensor protein AMPK. We demonstrated that simvastatin-induced G0/G1 cell cycle arrest was regulated by AMPK activation and STAT3 inactivation to transcriptionally increase p21 expression and stabilize p27 protein expression by inhibiting Skp2 expression. In addition, we also showed that this effect could be recovered by treatment with mevalonate, the product of HMG-CoA reductase. In contrast, in a xenograft animal model, we found that simvastatin treatment significantly suppressed HepG2 tumor growth and tumor weight. The results of our tumor section evaluations by IHC showed that simvastatin treatment increased p21 and p27 expression and AMPK activation and decreased Skp2 expression and STAT3 phosphorylation. These data provide evidence of a novel mechanism explaining the beneficial anticancer effects of simvastatin. These results are summarized in Figure 7e.
AMPK has been shown to promote accumulation of p53, which upregulates the protein expression of p21 at the transcriptional level and phosphorylates p27 at T198 to increase protein stability, thereby causing G0/G1 phase arrest.39, 40, 41, 42 In our experiments, we observed that simvastatin activated AMPK and induced p21 and p27 expression to cause G0/G1 phase arrest in HepG2 (p53 wild-type) and Hep3B (p53 mutant) cells (Figures 1a and d). We also found that simvastatin-induced p21 expression at the transcriptional level and promoted p27 accumulation by preventing proteasomal degradation (Figure 2). However, genetic knockdown of AMPK did not reduce p27 protein expression levels in our study (Figure 3c). These results suggested that simvastatin may regulate p21 gene expression induced by AMPK activation via other transcriptional machinery that functions independently of p53 activity such as FoxO1- and/or FoxO3-mediated p21 mRNA transcription in neonatal cardiomyocytes.43 Moreover, the above results suggest that simvastatin may not promote p27 protein accumulation through AMPK-mediated p27 phosphorylation at T198.
The p27 stability could be regulated by Skp2 and certain ubiquitin proteins.30 Previous studies have suggested that targeting Skp2 results in p27-mediated G0/G1 phase arrest.43 These findings indicate that Skp2 was crucial for regulating p27 expression in our models. Consistent with this finding, we observed that simvastatin decreased Skp2 expression at both the mRNA and the protein level concurrently with increases in p27 accumulation, resulting in G0/G1 phase arrest (Figures 4a and b). Skp2 overexpression reduced p27 protein levels and inhibited G0/G1 phase arrest in simvastatin-treated HepG2 cells (Figure 4d). The functions of Skp2 have been investigated, and previous studies have demonstrated that its expression can be regulated at the transcriptional level, as can its cell cycle-dependent degradation.32, 44, 45 In our study, we found that simvastatin decreased Skp2 expression mainly at the transcriptional level (Figure 4b) and that this effect may be mediated by STAT3 inhibition in HepG2 cells. Constitutive STAT3 activation maintained much higher levels of Skp2 and lower levels of p27 and reduced the G0/G1 phase cell population in simvastatin-treated HepG2 cells (Figures 5b and d). These results suggest that simvastatin inhibited the STAT3/Skp2 axis to promote p27 accumulation, resulting in G0/G1 phase arrest. Previous studies have shown that several mediators, including ERK1/2, E2F1, Akt, PPARγ, FOXP3 and STAT3, regulate Skp2 expression at the transcriptional level.35, 46, 47, 48, 49, 50 Recent studies have also reported that simvastatin can inhibit ERK1/2, Akt and STAT3 expression to suppress tumor cell growth and induce cell apoptosis.51, 52 However, no reports regarding STAT3 and Skp2 regulation in simvastatin-induced growth arrest in HCC are available. In this study, we demonstrated that simvastatin suppressed HCC cell growth by reducing Skp2 expression to cause p27 accumulation and induced G0/G1 phase arrest via STAT3 inhibition. It is possible that simvastatin may act through other pathways, such as ERK1/2 and Akt, to decrease Skp2 expression. These ideas will be evaluated in the future.
Mevalonate is synthesized by HMG-CoA reductase, and mevalonate pathway dysregulation promotes oncogenic transformation.53 Statins are well-known HMG-CoA reductase inhibitors and have been reported to inhibit renal cancer cell growth and metastasis.52 A recent report also suggested that statins inhibit IL-6-induced STAT3 phosphorylation and that this effect can be reversed by mevalonate in human hepatocytes.54 However, no studies demonstrating that maintaining mevalonate levels can facilitate the maintenance of much higher STAT3/Skp2 activity levels and lower p27 levels to prevent G0/G1 phase arrest in statins-treated cells are available. In our study, we found that mevalonate treatment could recover STAT3 and Skp2 expression, reduce AMPK phosphorylation, and downregulate p27- and p21-mediated G0/G1 arrest in simvastatin-treated HepG2 cells (Figures 6a and b). Studies focusing specifically on AMPK have shown that some mevalonate downstream products are essential for cell proliferation and survival. Moreover, these studies have also indicated that the anticancer effects of statins on AMPK activation may be mediated, at least in part, through inhibition of this pathway.23 The mechanisms by which mevalonate abrogated statin-induced AMPK activation are important and need to be evaluated in future studies.
In this study, we demonstrated that simvastatin-induced p27- and p21-mediated growth arrest by inhibiting STAT3/Skp2 and activating AMPK in HCC cells. Consistent with this finding, we also observed that simvastatin treatment inhibited tumor growth and reduced tumor volume in a mouse xenograft model of human HepG2 tumors. Tumor tissues evaluated using IHC assay also showed that simvastatin treatment increased p21 and p27 expression, increased AMPK activation, decreased Skp2 expression and reduced STAT3 phosphorylation. Furthermore, previous clinical studies have reported that statin use is associated with relative reductions in the risks of colorectal cancer, breast cancer and prostate cancer.55, 56 These findings suggest that statins warrant further investigation in chemoprevention and therapeutic clinical trials. In our study population, patients were newly diagnosed with HCC and had undergone liver resection as their initial HCC therapy. In addition, these patients must have used statins for >80 days (Supplementary Figure S4). Data regarding the cumulative incidence of overall mortality are shown in Supplementary Figure S3. These data suggest that statin use was associated with significantly lower overall mortality in HCC patients after liver resection than statin non-use. Therefore, statin use as a clinical therapy in HCC patients may be warranted. The findings of this study serve as evidence of the possible usefulness of statins in HCC patients and suggest that statin use has clinical significance, as statins can reduce the incidence of tumor recurrence, enhance chemoprevention and increase overall survival.
Materials and methods
Reagents and antibodies
Simvastatin was purchased from Cayman (Ann Arbor, MI, USA) and propidium iodide (PI), CHX, MG132 and mevalonate were obtained from Sigma (St. Louis, MO, USA). TRIzol reagent was obtained from Invitrogen (Carlsbad, CA, USA). Antibodies specific to p21, p27, cyclin D1, cyclin E1, phospho-AMPK, AMPK, phospho-STAT3 and STAT3 were purchased from Cell Signaling Technology (Danvers, MA, USA), the antibody specific to p45SKP2 was purchased from Invitrogen, and the antibody specific to β-actin was purchased from Santa Cruz (Santa Cruz, CA, USA).
Cell culture
Human hepatoma cell lines, HepG2 and Hep3B were cultured in Dulbecco’s modified Eagles’ medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 1% antibiotic solution (Invitrogen). Each cell line was maintained at 37 °C in a 5% CO2 incubator.
Cell viability and viable cell counts
Cell viability was determined using a Cell Counting Kit-8 Assay Kit (Sigma). HepG2 and Hep3B cells were seeded into 96-well plates at a density of 1 × 104 cells per well and were treated with the indicated concentrations (0–40 μg/ml) of simvastatin for 24 or 48 h. After the cells had incubated, we assessed their viability using the above assay kit, in accordance with the manufacturer’s instructions. The absorbance was detected at 450 nm with an ELISA Plate Reader (PerkinElmer, Waltham, MA, USA). To measure and calculate the fractions of dead and live cells, we determined the counts of death cells by Trypan blue exclusion in a hemocytometer. The results were expressed as a percentage of the control.
DNA content assay
Cell cycle distributions were determined by DNA content assay. HepG2 and Hep3B cells were seeded into six-well plates and treated with the indicated concentrations (0-20 μg/ml) of simvastatin for 48 h. At incubation time, the cells were harvested and fixed in 70% ethanol at 4 °C overnight, and then they were washed with phosphate-buffered saline (PBS) and stained with 20 μg/ml PI at 37 °C for 30 min. The cell populations were analyzed by FAC Sort flow cytometry (BD Biosciences, San Jose, CA, USA).
Reverse transcription polymerase chain reaction (RT-PCR) and real-time PCR
In all, 1μg of total RNA from the culture cells was extracted using Trizol Reagent (Invitrogen). Complementary DNA (cDNA) synthesized with the transcriptor first strand cDNA synthesis kit (Clontech, Mountain View, CA, USA) according to the manufacturer’s instructions. To amplify the target genes, PCR was performed using specific primers: p21, forward 5′-GAGCGATGGAACTTCGACTT-3′ and reverse 5′-GGCGTTTGGAGTGGTAGAAA-3′; p27, forward 5′-TCTACTGCGTGGCTTGTCAG-3′ and reverse 5′-CTGTATTTGGAGGCACAGCA-3′; Skp2, forward 5′-TGAGCTGCTCTTGGGAATCT-3′ and reverse 5′-GTCTGGGACAGCTGCTTAGG-3′; GAPDH as endogenous control, forward 5′-ACCACAGTCCATGCCATCAC-3′ and reverse 5′-TCCACCACCCTGTTGCTGTA-3′. PCR amplification used TEMPase Hot Start DNA Polymerase (Ampliqon, Hamburg, Germany). Procedures were described as follows: 50 ng of cDNA was added to 20 μl of PCR mixture. The PCR mixture was performed after 15 min of denaturation at 95 °C and then amplification for 25 cycles contained the 15 s of denaturation at 95 °C, 30 s of annealing step at 60 °C, 60 s of extension at 72 °C, and another 10 min at 72 °C after the last cycle. The PCR products were separated on 2% agarose gels. For real-time PCR analysis, the synthesized cDNAs were mixed with 2X SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) were detected by the StepOnePlus Real-Time PCR System (Applied Biosystems). A pair of gene-specific forward and reverse primers (p21: 5′-AGACTCTCAGGGTCGAAAAC-3′, 5′-TGGAGTGGTAGAAATCTGTCATG-3′; p27: 5′-TGCAACCGACGATTCTTCTAC-3′, 5′-CTTCTGTTCTGTTGGCTCTTTTG-3′; Skp2: 5′-CTGTCTCAAGGGGTGATTGC-3′, 5′-TTCGATAGGTCCATGTGCTG-3′; GAPDH: 5′-ACCACAGTCCATGCATCAC-3′; 5′-TCCACCACCCTGTTGCTGT-3′). All reactions were performed in triplicate. The relative amounts of mRNAs were calculated using the comparative CT method. Human GAPDH mRNA was used as the internal control.
siRNA transfections
Human AMPK (siRNAs; Santa Cruz), p21, p27 and control siRNAs (GE Dharmacon, Lafayette, CO, USA) were transiently transfected into cells with INTERFERin transfection reagent according to the manufacturer's instructions (Polyplus Transfection, New York, NY, USA). After 24 h, the cells were treated with simvastatin (0 or 10 μg/ml) for 48 h and harvested for immunoblotting or DNA content assay.
Plasmid DNA transfections
293T cells were plated at a density of 1 × 105 cells per well in six-well plates overnight. The cells were transfected using JetPEI transfection reagent (Polyplus Transfection), according to the retrovirus manufacturer’s instructions, with a lenti- or retroviral vector encoding constitutively activated STAT3-GFP (EF.STAT3C.Ubc.GFP, Addgene plasmid 24983, Addgene, Cambridge, MA, USA)57 or p-Babe-N-tag-SKP258 and expression vectors encoding the packing proteins gag-pol and VSV-G. Viral supernatants were collected starting 48 and 96 h after transfection and filtered through a 0.45-μm filter. Then, for viral transduction processing, we added HepG2 cells to infecting medium with 8 μg/ml polybrene for 48 h. The HepG2 cells were subsequently treated with simvastatin (0 or 20 μg/ml) for 48 h and harvested for immunoblotting or cell cycle analysis.
Luciferase assay
Cells were co-transfected with a pGL4.18-SKP2 promoter reporter plasmid58 and a control Renilla luciferase reporter plasmid (Promega, Madison, WI, USA) for 24 h. At the indicated time, the cells were treated with simvastatin 20 μg/ml for 24 h and then harvested. Luciferase activity was determined using a dual-luciferase reporter assay system (Promega). Light units were normalized to Renilla luciferase activity.
Immunoblotting
After the indicated treatments, cells were harvested and lysed in PRO-PREP protein extraction solution (iNtRON, Taipei, Taiwan) containing a protease inhibitor cocktail. Cell lysates were centrifuged at 12 000 g for 15 min at 4 °C, and the supernatants were collected. The protein concentration of the samples was measured using a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, USA). Then, 40–50 μg protein from each sample was separated on a 10% or 12% SDS-polyacrylamide gel before being transferred onto equilibrated polyvinylidene difluoride membranes. After being blocked, the membranes were incubated with the appropriate primary antibodies at 4 °C overnight. Then, membranes were incubated with the appropriate secondary antibodies at 4 °C for 2 h, and the signals were detected using an Odyssey Imaging System (Odyssey, Lincoln, NE, USA). β-Actin was used as a loading control in the immunoblotting analysis.
Tumor growth in the xenograft mouse model
Male BALB/c nude mice (6–8 weeks old) were purchased from the National Laboratory Animal Center (NLAC, Taipei, Taiwan). HepG2 cells were cultured with DMEM supplemented with 10% FBS. Viable HepG2 cells (1.0 × 107 cells in 0.2 ml of serum-free DMEM) were injected subcutaneously into the upper right flanks of the mice. Treatment was initiated when tumor volumes reached a mean size of approximately 100 mm3. The mice were subsequently subjected to intraperitoneal injections of saline or simvastatin (20 mg/kg body weight) twice a day for 2 weeks. Tumor volumes were measured every other day with calipers and calculated using the following formula: largest diameter × (smallest diameter)2 × 0.5. After being treated for 14 days, the mice were killed. The tumors were collected, and the weight of each tumor was measured. All animal care and experimental procedures were approved and conducted by the Committee for Animal Experiments, National Chung Hsing University, Taichung, Taiwan (approved document La-1051401).
Histological analysis and IHC
Tumor tissues from the HepG2 tumor-bearing control mice and simvastatin treatment mice were harvested, fixed in 10% formalin and embedded in paraffin. The paraffin sections were subsequently stained with hematoxylin and eosin for morphological observation. For IHC, the paraffin slides were deparaffinized and rehydrated in xylene and ethanol. Antigen retrieval was performed in Tris/EDTA (pH 9.0) buffer by heating for 30 min, followed by incubation in an enzymatic antigen retrieval solution for 10 mm for 20 min. The slides were incubated with 0.3% H2O2 in TBS for 15 min and washed with 0.025% Triton X-100 in TBS and then blocked with 1% BSA in TBS for 2 h at room temperature before being incubated at 4 °C overnight with the appropriate primary antibody. Then, the slides were incubated with the appropriate HRP-conjugated secondary antibody for 1 h at room temperature and visualized using 3,3' diaminobenzidine substrate and counterstaining with hematoxylin. Images of the slides were acquired using inverted microscopy.
Statistical analysis
All assays were performed as three independent experiments in duplicate or triplicate. Data were analyzed using Student’s t-test, and significant differences were inferred at a P-value of 0.05.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D et al. Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90.
El-Serag HB, Rudolph KL . Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 2007; 132: 2557–2576.
El-Serag HB . Hepatocellular carcinoma. N Engl J Med 2011; 365: 1118–1127.
Pang RW, Poon RT . From molecular biology to targeted therapies for hepatocellular carcinoma: the future is now. Oncology 2007; 72 (Suppl 1): 30–44.
Wu CY, Chen YJ, Ho HJ, Hsu YC, Kuo KN, Wu MS et al. Association between nucleoside analogues and risk of hepatitis B virus-related hepatocellular carcinoma recurrence following liver resection. JAMA 2012; 308: 1906–1914.
Hsu YC, Wu CY, Lin JT, Ho HJ, Su CW, Lee TY et al. Association of nucleos(t)ide analogue therapy with reduced risk of hepatocellular carcinoma in patients with chronic hepatitis B: a nationwide cohort study. Gastroenterology 2014; 147: 143–151 e5.
Lee TY, Lin JT, Zeng YS, Chen YJ, Wu MS, Wu CY et al. Association between nucleos(t)ide analogue and tumor recurrence in HBV-related hepatocellular carcinoma after radiofrequency ablation. Hepatology 2016; 63: 1517–1527.
Chen HP, Shieh JJ, Chang CC, Chen TT, Lin JT, Wu MS et al. Metformin decreases hepatocellular carcinoma risk in a dose-dependent manner: population-based and in vitro studies. Gut 2013; 62: 606–615.
Relja B, Meder F, Wilhelm K, Henrich D, Marzi I, Lehnert M et al. Simvastatin inhibits cell growth and induces apoptosis and G0/G1 cell cycle arrest in hepatic cancer cells. Int J Mol Med 2010; 26: 735–741.
Lonardo A, Loria P . Potential for statins in the chemoprevention and management of hepatocellular carcinoma. J Gastroenterol Hepatol 2012; 27: 1654–1664.
Tournier B, Chapusot C, Courcet E, Martin L, Lepage C, Faivre J et al. Why do results conflict regarding the prognostic value of the methylation status in colon cancers? The role of the preservation method. BMC Cancer 2012; 12: 12.
Roy M, Kung HJ, Ghosh PM . Statins and prostate cancer: role of cholesterol inhibition vs. prevention of small GTP-binding proteins. Am J Cancer Res 2011; 1: 542–561.
Bardou M, Barkun A, Martel M . Effect of statin therapy on colorectal cancer. Gut 2010; 59: 1572–1585.
Lai SW, Liao KF, Lai HC, Muo CH, Sung FC, Chen PC et al. Statin use and risk of hepatocellular carcinoma. Eur J Epidemiol 2013; 28: 485–492.
Ghosh-Choudhury N, Mandal CC, Ghosh Choudhury G . Simvastatin induces derepression of PTEN expression via NFkappaB to inhibit breast cancer cell growth. Cell Signal 2011; 22: 749–758.
Kochuparambil ST, Al-Husein B, Goc A, Soliman S, Somanath PR . Anticancer efficacy of simvastatin on prostate cancer cells and tumor xenografts is associated with inhibition of Akt and reduced prostate-specific antigen expression. J Pharmacol Exp Ther 2011; 336: 496–505.
Choi HC, Song P, Xie Z, Wu Y, Xu J, Zhang M et al. Reactive nitrogen species is required for the activation of the AMP-activated protein kinase by statin in vivo. J Biol Chem 2008; 283: 20186–20197.
Misirkic M, Janjetovic K, Vucicevic L, Tovilovic G, Ristic B, Vilimanovich U et al. Inhibition of AMPK-dependent autophagy enhances in vitro antiglioma effect of simvastatin. Pharmacol Res 2012; 65: 111–119.
Kou R, Sartoretto J, Michel T . Regulation of Rac1 by simvastatin in endothelial cells: differential roles of AMP-activated protein kinase and calmodulin-dependent kinase kinase-beta. J Biol Chem 2009; 284: 14734–14743.
Sun W, Lee TS, Zhu M, Gu C, Wang Y, Zhu Y et al. Statins activate AMP-activated protein kinase in vitro and in vivo. Circulation 2006; 114: 2655–2662.
Yang PM, Liu YL, Lin YC, Shun CT, Wu MS, Chen CC et al. Inhibition of autophagy enhances anticancer effects of atorvastatin in digestive malignancies. Cancer Res 2010; 70: 7699–7709.
Rao S, Porter DC, Chen X, Herliczek T, Lowe M, Keyomarsi K et al. Lovastatin-mediated G1 arrest is through inhibition of the proteasome, independent of hydroxymethyl glutaryl-CoA reductase. Proc Natl Acad Sci USA 1999; 96: 7797–7802.
Motoshima H, Goldstein BJ, Igata M, Araki E . AMPK and cell proliferation—AMPK as a therapeutic target for atherosclerosis and cancer. J Physiol 2006; 574: 63–71.
Sala SG, Munoz U, Bartolome F, Bermejo F, Martin-Requero A . HMG-CoA reductase inhibitor simvastatin inhibits cell cycle progression at the G1/S checkpoint in immortalized lymphocytes from Alzheimer's disease patients independently of cholesterol-lowering effects. J Pharmacol Exp Ther 2008; 324: 352–359.
Subramaniam A, Shanmugam MK, Perumal E, Li F, Nachiyappan A, Dai X et al. Potential role of signal transducer and activator of transcription (STAT)3 signaling pathway in inflammation, survival, proliferation and invasion of hepatocellular carcinoma. Biochim Biophys Acta 2013; 1835: 46–60.
Sethi G, Chatterjee S, Rajendran P, Li F, Shanmugam MK, Wong KF et al. Inhibition of STAT3 dimerization and acetylation by garcinol suppresses the growth of human hepatocellular carcinoma in vitro and in vivo. Mol Cancer 2014; 13: 66.
Sun X, Zhang J, Wang L, Tian Z . Growth inhibition of human hepatocellular carcinoma cells by blocking STAT3 activation with decoy-ODN. Cancer Lett 2008; 262: 201–213.
Sutter AP, Maaser K, Hopfner M, Huether A, Schuppan D, Scherubl H et al. Cell cycle arrest and apoptosis induction in hepatocellular carcinoma cells by HMG-CoA reductase inhibitors. Synergistic antiproliferative action with ligands of the peripheral benzodiazepine receptor. J Hepatol 2005; 43: 808–816.
Lee SK, Kim YS . Phosphorylation of eIF2alpha attenuates statin-induced apoptosis by inhibiting the stabilization and translocation of p53 to the mitochondria. Int J Oncol 2013; 42: 810–816.
Carrano AC, Eytan E, Hershko A, Pagano M . SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol 1999; 1: 193–199.
Lu Z, Hunter T . Ubiquitylation and proteasomal degradation of the p21(Cip1), p27(Kip1) and p57(Kip2) CDK inhibitors. Cell Cycle 2010; 9: 2342–2352.
Kossatz U, Dietrich N, Zender L, Buer J, Manns MP, Malek NP et al. Skp2-dependent degradation of p27kip1 is essential for cell cycle progression. Genes Dev 2004; 18: 2602–2607.
Wu L, Grigoryan AV, Li Y, Hao B, Pagano M, Cardozo TJ et al. Specific small molecule inhibitors of Skp2-mediated p27 degradation. Chem Biol 2012; 19: 1515–1524.
Huang H, Zhao W, Yang D . Stat3 induces oncogenic Skp2 expression in human cervical carcinoma cells. Biochem Biophys Res Commun 2012; 418: 186–190.
Wei Z, Jiang X, Qiao H, Zhai B, Zhang L, Zhang Q et al. STAT3 interacts with Skp2/p27/p21 pathway to regulate the motility and invasion of gastric cancer cells. Cell Signal 2013; 25: 931–938.
Maciejak A, Leszczynska A, Warchol I, Gora M, Kaminska J, Plochocka D et al. The effects of statins on the mevalonic acid pathway in recombinant yeast strains expressing human HMG-CoA reductase. BMC Biotechnol 2013; 13: 68.
Gopalan A, Yu W, Sanders BG, Kline K . Simvastatin inhibition of mevalonate pathway induces apoptosis in human breast cancer cells via activation of JNK/CHOP/DR5 signaling pathway. Cancer Lett 2013; 329: 9–16.
Buhaescu I, Izzedine H . Mevalonate pathway: a review of clinical and therapeutical implications. Clin Biochem 2007; 40: 575–584.
Schiappacassi M, Lovisa S, Lovat F, Fabris L, Colombatti A, Belletti B et al. Role of T198 modification in the regulation of p27(Kip1) protein stability and function. PLoS ONE 2011; 6: e17673.
Imamura K, Ogura T, Kishimoto A, Kamonishi M, Esumi H . Cell cycle regulation via p53 phosphorylation by a 5'-AMP activated protein kinase activator, 5-aminoimidazole- 4-carboxamide-1-beta-D-ribofuranoside, in a human hepatocellular carcinoma cell line. Biochem Biophys Res Commun 2001; 287: 562–567.
Rattan R, Giri S, Singh AK, Singh I . 5-Aminoimidazole-4-carboxamide-1-beta-d-ribofuranoside inhibits cancer cell proliferation in vitro and in vivo via AMP-activated protein kinase. J Biol Chem 2005; 280: 39582–39593.
Ikeda Y, Sato K, Pimentel DR, Sam F, Shaw RJ, Dyck JRB et al. Cardiac-specific deletion of LKB1 leads to hypertrophy and dysfunction. J Biol Chem 2009; 284: 35839–35849.
Chen Q, Xie W, Kuhn DJ, Voorhees Q, Lopez-Girona Q, Mendy Q et al. Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood 2008; 111: 4690–4699.
Wei W, Ayad NG, Wan Y, Zhang GJ, Kirschner MW, Kaelin WG et al. Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature 2004; 428: 194–198.
Frescas D, Pagano M . Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer 2008; 8: 438–449.
Villanueva J, Yung Y, Walker JL, Assoian RK . ERK activity and G1 phase progression: identifying dispensable versus essential activities and primary versus secondary targets. Mol Biol Cell 2007; 18: 1457–1463.
Zhang L, Wang C . F-box protein Skp2: a novel transcriptional target of E2F. Oncogene 2006; 25: 2615–2627.
Reichert M, Saur D, Hamacher R, Schmid RM, Schneider G . Phosphoinositide-3-kinase signaling controls S-phase kinase-associated protein 2 transcription via E2F1 in pancreatic ductal adenocarcinoma cells. Cancer Res 2007; 67: 4149–4156.
Meng J, Ding Y, Shen A, Yan M, He F, Ji H et al. Overexpression of PPARgamma can down-regulate Skp2 expression in MDA-MB-231 breast tumor cells. Mol Cell Biochem 2010; 345: 171–180.
Zuo T, Liu R, Zhang H, Chang X, Liu Y, Wang L et al. FOXP3 is a novel transcriptional repressor for the breast cancer oncogene SKP2. J Clin Invest 2007; 117: 3765–3773.
Yanae M, Tsubaki M, Satou T, Itoh T, Imano M, Yamazoe Y et al. Statin-induced apoptosis via the suppression of ERK1/2 and Akt activation by inhibition of the geranylgeranyl-pyrophosphate biosynthesis in glioblastoma. J Exp Clin Cancer Res 2010; 30: 74.
Fang Z, Tang Y, Fang J, Zhou Z, Xing Z, Guo Z et al. Simvastatin inhibits renal cancer cell growth and metastasis via AKT/mTOR, ERK and JAK2/STAT3 pathway. PLoS ONE 2013; 8: e62823.
Clendening JW, Pandyra A, Boutros PC, Ghamrasni EI, Khosravi F, Trentin GA et al. Dysregulation of the mevalonate pathway promotes transformation. Proc Natl Acad Sci USA 2010; 107: 15051–15056.
Arnaud C, Burger F, Steffens S, Veillard NR, Nguyen TH, Trono D et al. Statins reduce interleukin-6-induced C-reactive protein in human hepatocytes: new evidence for direct antiinflammatory effects of statins. Arterioscler Thromb Vasc Biol 2005; 25: 1231–1236.
Poynter JN, Gruber SB, Higgins PD, Almog R, Bonner JD, Rennert HS et al. Statins and the risk of colorectal cancer. N Engl J Med 2005; 352: 2184–2192.
Hindler K, Cleeland CS, Rivera E, Collard CD . The role of statins in cancer therapy. Oncologist 2006; 11: 306–315.
Hillion J, Dhara S, Sumter TF, Mukherjee M, Di Cello F, Belton A et al. The high-mobility group A1a/signal transducer and activator of transcription-3 axis: an achilles heel for hematopoietic malignancies? Cancer Res 2008; 68: 10121–10127.
Hsieh HY, Shieh JJ, Chen CJ, Pan MY, Yang SY, Lin SC et al. Prodigiosin down-regulates SKP2 to induce p27(KIP1) stabilization and antiproliferation in human lung adenocarcinoma cells. Br J Pharmacol 2012; 166: 2095–2108.
Acknowledgements
This research was supported in part by Taichung Veterans General Hospital (TCVGH-1033304C) and Taiwan National Health Research Institute (PH-104-PP-44, PH-104-PP-45). We are grateful to Dr. Chia-Che Chang for kindly providing p-Babe-N-tag-SKP2 and pGL4.18-SKP2 promoter reporter plasmid. We also thank Dr. Linzhao Cheng, who kindly provided the EF.STAT3C.Ubc.GFP plasmid. The technical support provided by Instrument Center of Department of Medical Research of Taichung Veterans General Hospital is gratefully acknowledge.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Stephanou
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Wang, ST., Ho, H., Lin, JT. et al. Simvastatin-induced cell cycle arrest through inhibition of STAT3/SKP2 axis and activation of AMPK to promote p27 and p21 accumulation in hepatocellular carcinoma cells. Cell Death Dis 8, e2626 (2017). https://doi.org/10.1038/cddis.2016.472
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2016.472
This article is cited by
-
O-GlcNAcylation of E3 ubiquitin ligase SKP2 promotes hepatocellular carcinoma proliferation
Oncogene (2024)
-
Dysregulated cholesterol regulatory genes in hepatocellular carcinoma
European Journal of Medical Research (2023)
-
STAT3 potentiates RNA polymerase I-directed transcription and tumor growth by activating RPA34 expression
British Journal of Cancer (2023)
-
Enhanced therapeutic efficacy of doxorubicin/cyclophosphamide in combination with pitavastatin or simvastatin against breast cancer cells
Medical Oncology (2023)
-
Increased Pro-Apoptotic and Anti-Proliferative Activities of Simvastatin Encapsulated PCL-PEG Nanoparticles on Human Breast Cancer Adenocarcinoma Cells
Journal of Cluster Science (2023)