Abstract
Sponges of the porifera family harbor some of the evolutionary most ancient orthologs of the B-cell lymphoma-2 (Bcl-2) family, a protein family critical to regulation of apoptosis. The genome of the sponge Geodia cydonium contains the putative pro-survival Bcl-2 homolog BHP2, which protects sponge tissue as well as mammalian Hek-293 and NIH-3T3 cells against diverse apoptotic stimuli. The Lake Baikal demosponge Lubomirskia baicalensis has been shown to encode both putative pro-survival Bcl-2 (LB-Bcl-2) and pro-apoptotic Bcl-2 members (LB-Bak-2), which have been implied in axis formation (branches) in L. baicalensis. However, the molecular mechanism of action of sponge-encoded orthologs of Bcl-2 remains to be clarified. Here, we report that the pro-survival Bcl-2 ortholog BHP2 from G. cydonium is able to bind the BH3 motif of a pro-apoptotic Bcl-2 protein, LB-Bak-2 of the sponge L. baicalensis. Furthermore, we determined the crystal structure of BHP2 bound to LB-Bak-2, which revealed that using a binding groove conserved across all pro-survival Bcl-2 proteins, BHP2 binds multi-motif Bax-like proteins through their BH3-binding regions. However, BHP2 discriminates against BH3-only bearing proteins by blocking access to a hydrophobic pocket that is critical for BH3 motif binding in pro-survival Bcl-2 proteins from higher organisms. This differential binding mode is reflected in a structure-based phylogenetic comparison of BHP2 with other Bcl-2 family members, which revealed that BHP2 does not cluster with either Bcl-2 members of higher organisms or pathogen-encoded homologs, and assumes a discrete position. Our findings suggest that the molecular machinery and mechanisms for executing Bcl-2-mediated apoptosis as observed in mammals are evolutionary ancient, with early regulation of apoptotic machineries closely resembling their modern counterparts in mammals rather than Caenorhabditis elegans or drosophila.
Similar content being viewed by others
Main
Sponges (Porifera) and Hydra (Cnidaria) are thought to be among the earliest metazoans, multicellular organisms that have differentiated tissues, yet they have evolved highly sophisticated mechanisms of homeostatic regulation.1 Sponge cells harbor a capacity for unlimited proliferation and differentiation potency,2 and as such a finely tuned control system necessary to maintain cellular homeostasis. Sequence analysis identified in the sponge orthologous genes to those in higher organisms that regulate apoptosis (programmed cell death).3
A critical group of proteins that regulate apoptosis are the B-cell lymphoma-2 (Bcl-2) family and these proteins4 regulate intrinsic (mitochondrial-mediated) apoptosis.5 The Bcl-2 family is divided into pro-survival and pro-apoptotic members, all of which share one or more of four conserved Bcl-2 homology (BH) motifs.6 Pro-survival Bcl-2 proteins including Bcl-2, Bcl-xL, Bcl-w, Mcl-1, A1 and Bcl-B block apoptosis, whereas the pro-apoptotic Bcl-2 members are necessary for initiation of cell death. The pro-apoptotic Bcl-2 proteins are further subdivided into two groups, those that only harbor a BH3 domain, which are termed the BH3-only proteins,7 and the multi-BH motif proteins Bax, Bak and Bok8 essential for cell death initiation. The BH3-only proteins including Bim, Puma, Bad and Noxa either directly activate pro-apoptotic Bax and Bak, or neutralize pro-survival Bcl-2 members by engaging their receptor binding groove using the α-helical BH3 domains.9 Upon activation, Bax and Bak oligomerize to cause organellar damage including permeabillization of the outer mitochondrial membrane,10, 11 thus releasing critical pro-apoptogenic factors such as cytochrome c and SMAC/Diablo from mitochondria activates caspases that destroy the cell.12 Detailed structural and binding studies have been performed on mammalian and worm apoptosis regulators showing many aspects of the mechanism of apoptosis are conserved across phyla.13
Evolutionary ancient ancestors of Bcl-2 proteins have been identified in sponges.14 The sponge Geodia cydonium expresses a putative pro-survival Bcl-2 homolog, BHP2, that protects sponge tissue against apoptosis induced by heat-shock or tributylin (an organic tin derivative) exposure, as well as mammalian Hek-293 and NIH-3T3 cells against serum withdrawal or tributylin.15 Similarly, the Lake Baikal demosponge Lubomirskia baicalensis harbors both putative pro-survival Bcl-2 (LB-Bcl-2) and pro-apoptotic Bcl-2 members (LB-Bak-2)16 that were postulated to be involved in axis formation (branches) in L. baicalensis, however their mechanism of action remains to be clarified. These findings indicate that the sponges maintain an apoptotic machinery to regulate homeostasis. To elucidate molecular mechanisms of apoptosis in these early lifeforms we have performed structural and binding studies on BHP2 and LB-Bak-2 from G. cydonium and L. baicalensis, respectively. We demonstrate that BHP2 shares the Bcl-2 fold with its higher organisms and, like them, bears a binding groove that binds the BH3-region of LB-Bak-2. Thus, mechanistically the sponge appears to replicate features of the mammalian rather than worm or fly apoptotic program and implies that the pro-apoptotic Bax-like proteins are regulated by interaction with the pro-survival Bcl-2 proteins in the sponge.
Results
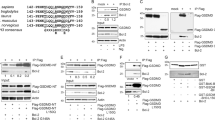
To resolve the molecular mechanism underlying Bcl-2-mediated apoptosis in sponges, we examined the ability of the pro-survival Bcl-2 homolog BHP2 from G. cydonium to bind pro-apoptotic Bcl-2 proteins. Biochemical studies of a related pro-survival Bcl-2 protein from the demosponge L. baicalensis were also considered, however lack of expression ultimately precluded an analysis (data not shown). As pro-apoptotic Bcl-2 orthologs have not been identified in G. cydonium, we determined the capacity of BHP2 to bind a Bak-2 homolog from a related demosponge L. baicalensis, LB-Bak-2, or indeed any other human pro-apoptotic Bcl-2 family member. Using recombinant BHP2 we measured binding to peptides spanning the BH3 motif of LB-Bak-2, as well as all human BH3-only proteins and Bak, Bax and Bok. BHP2 bound LB-Bak-2 BH3 peptides with high affinity (KD=65 nM) (Figure 1a), whereas with the exception of Hrk (KD=3760 nM) (Figure 1b), no other BH3 motif of human pro-apoptotic Bcl-2 proteins displayed any significant binding (Figure 1c).
BHP2 interactions with BH3 domain peptides of pro-apoptotic Bcl-2 proteins. The affinity of recombinant BHP2 for BH3 domain peptides (26-mers, except for a Bax 28-mer and a Bid 34-mer) was assessed using isothermal titration calorimetry. KD values (in nM) are the means of three experiments±S.D. NB, no binding detected, N denotes stoichiometry
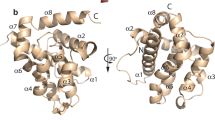
The structural basis for BH3 motif binding by BHP2, and its ability to discriminate against human pro-apoptotic Bcl-2 proteins over their sponge counterparts, was determined by solving the crystal structure of BHP2 bound to the LB-Bak-2 BH3 motif (Figure 2; Table 1). Like other Bcl-2 orthologs BHP2 adopts a globular helical bundle fold comprising 8 α-helices, with helices 2–5 forming the canonical binding groove observed in all other pro-survival Bcl-2 family members.6 Clear and continuous electron density is observed for residues 18–38 and 67–198, with the intervening linker region being largely disordered except for a short helical turn formed by residues 53–57 that anchor the loop to the solvent exposed side of helix 6 from the main globular motif (Figure 3). A DALI17 similarity analysis revealed that among all Bcl-2 proteins, complexes of Bcl-xL (PDB ID 1PQ1) and Bcl-2 (PDB ID 5FCG) bound to BH3 domain peptides are most similar to BHP2, with r.m.s.d. values of 2.2 Å in both cases over 137 and 134 Cα atoms, respectively.
Crystal structure of BHP2 bound to LB-Bak-2 BH3 domain. (a) BHP2 (pink) in complex with the LB-Bak-2 BH3 domain (violet). BHP2 helices are labeled α1–8. The view in (a) is into the hydrophobic binding groove formed by helices α3–5. (b) Bcl-xL (cyan) in complex with the Bim BH3 domain (orange).43 The view is as in (a). (c) Vaccinia virus F1L (sand) in complex with the Bak BH3 domain (green).45 The view is as in (a). (d) CED9 (blue) in complex with the EGL1 BH3 domain (yellow).44 The view is as in (a).
Similar to pro-survival Bcl-2 proteins from higher organisms, BHP2 engages the LB-Bak-2 BH3 motif via the canonical Bcl-2 binding groove formed by α-helices 2–5. A total of 913 Å2 of solvent accessible surface is buried in the process. As expected, BHP2 Arg135BHP2 from the conserved BH1 motif forms an ionic interaction with the LB-Bak-2 Asp80Bak, an interaction conserved in almost all Bcl-2-BH3 motif interactions. Interestingly, Asp80Bak is also engaged in a second ionic interaction with His132BHP2, which in turn also contacts Asp83Bak, leading to the formation of a network of ionic interactions centering on LB-Bak-2 Asp80. In addition, the BHP2:LB-Bak-2 interface features several hydrogen bonds between Glu118BHP2 and Ser69Bak, Tyr190BHP2 and Asp83Bak, Arg135BHP2 and Ala76Bak, and Glu118BHP2 and Trp108BHP2 with Ser65Bak. A hallmark of pro-survival Bcl-2:pro-apoptotic BH3 motif interactions is the engagement of four highly conserved hydrophobic residues from the BH3 motif by the ligand binding groove of pro-survival Bcl-2.7 While BHP2 accommodates V71, L75 and V82, unexpectedly F78 is not captured by a hydrophobic pocket at the floor of the binding grove. Instead, the helical turn in LB-Bak-2 BH3 that features F78 is bulged out, allowing F78 to sit on top of BHP2 I94 (Figure 4a). This shift is enforced by the positioning of I94 in BHP2, where the protruding Ile side chain prevents access of LB-Bak-2 F78 into the typical position adopted in other pro-survival Bcl-2:BH3 motif complexes (Supplementary Figure S2).
(a) Detailed view of the BHP2:LB-Bak-2 BH3 interface. The BHP2 surface, backbone and binding groove are shown in gray and pink respectively, whereas LB-Bak-2 BH3 is shown in violet. The four key hydrophobic residues of LB-Bak-2 (V71, L75, F78 and V82) on protruding into the binding groove, and the conserved salt bridge formed by LB-Bak D80 and BHP2 R135 are labeled, as well as residues involved in hydrogen bonds. Comparison of the P3 pocket among pro-survival Bcl-2: pro-apoptotic BH3 domain complexes. (b) BHP2:LB-Bak-2 complex. (c) Bcl-xL:Bim complex. (d) CED9:EGL1 complex. (e) F1L:Bak complex
To further understand the evolutionary relationship between BHP2 and Bcl-2 family proteins from higher organisms, we performed a structure-guided evolutionary analysis using pro-survival Bcl-2 family proteins of known structure by performing a pairwise comparison of experimentally determined structures of pro-survival Bcl-2 proteins using r.m.s.d. values for equivalent Cα atoms. We found that BHP2 did not cluster with other Bcl-2 proteins, with family members originating from higher organisms forming one cluster and pathogen-encoded family members forming a second cluster (Figure 5c).
Discussion
Bcl-2 proteins are the major arbiter of intrinsic apoptosis in higher organisms regulating the release of apoptogenic factors through the mitochondrial outer membrane.6 Essentially, there are two subgroups of the Bcl-2 family, the folded Bcl-2-like proteins which are either pro-survival (Bcl-2, Bcl-xL, Mcl-1, A1 and Bcl-B or pro-apoptotic (Bak, Bax, Bok, the latter two appear later in evolution)), and a phylogenetically distinct pro-apoptotic BH3-only family (Bim, Bad, Bmf, tBid, Bik, Noxa, Puma, Hrk, in mammals) that are unstructured.7 However, there are species-specific differences in their mode of action and it is not clear if the BH3-only proteins exist in all phyla.18 Despite the identification of ancient sequence orthologs of Bcl-2 family members in demosponges (Figures 5a and b)15, 16 and evidence that sponge Bcl-2 orthologs are able to inhibit apoptosis,15 it remained unclear if such ancient Bcl-2 proteins function mechanistically in a manner analogous to their modern counterparts.
BHP2 is able to protect HEK-293 cells against apoptosis under cell culture conditions,15 however, the molecular basis for this apoptosis inhibition in a human cell line remains to be understood. We find that BHP2 is able to bind the BH3 motif of the L. baicalensis pro-apoptotic Bcl-2 homolog LB-Bak-2 with high affinity. However, only low affinity to Hrk BH3 and no binding to other BH3 peptides from human Bcl-2 proteins including those from human Bak, Bax or BH3-only proteins was measured. Consequently, there is no obvious mechanism for BHP2 to inhibit intrinsic apoptosis in human cell lines. However, a similar situation was observed when human Bcl-2 was overexpressed in C. elegans.19, 20 Bcl-2 potently protected against apoptosis in the worm, despite its low affinity for pro-apoptotic Egl1 (KD=7.1 μM).21
The crystal structure of the BHP2:LB-Bak-2 BH3 complex demonstrates BHP2 utilizes the conserved canonical binding groove found in all other pro-survival Bcl-2 family members to date. Despite the overt mechanistic similarity with mammalian Bcl-2 proteins there are key differences in how BHP2 engages BH3 motifs. BHP2 engagement of the LB-Bak-2 BH3 motif relies only on the use of three conserved hydrophobic residues in the BH3 domain, with the third hydrophobic residue F78Bak being pushed out of the BHP2 ligand binding groove and accommodated via a pronounced bulge in the otherwise helical LB-Bak-2 BH3 motif. The unexpected absence of a pocket suitable to accommodate the LB-Bak-2 F78 residue is likely to be a major structural determinant for the selectivity of BHP2 for BH3 motif ligands. Indeed, of all human pro-apoptotic BH3 motifs tested, only Hrk revealed a modest affinity of 3.7 μM for BHP2, with the BH3 motif of human Bak displaying no detectable binding.
Interestingly, a major difference between LB-Bak-2 and human Bak BH3 motifs centers on LB-Bak-2 F78, the equivalent residue in human Bak is an isoleucine (I81) that is unable to rotate in the manner that allows the corresponding phenylalanine in LB-Bak-2 to swing out of the way to minimize steric hindrance. The observed differences in the mode of engagement of a BH3 motif with BHP2 compared with other pro-survival Bcl-2 proteins that utilize four pockets to bind BH3 motifs is also reflected in the structure-based phylogenetic analysis of BHP2 (Figure 5c), which suggests that BHP2 is unique when compared with mammalian- and pathogen- encoded pro-survival Bcl-2 proteins.
Our findings provide a mechanistic platform for understanding apoptosis regulation by early Bcl-2 orthologs. Apoptotic features in sponges were first observed in hibernating sponges and like higher metazoans, they have also been observed during developmental processes.2, 22 Furthermore, dysregulated apoptosis in these simple organisms is a potential mechanism for disease. Surprisingly, it has been observed that like humans, Hydra are susceptible to tumors where deregulated apoptosis appears to be the underlying cause.23 However, the molecular underpinnings of these observations have not yet been elucidated.
The presence of functional pro-survival Bcl-2 and pro-apoptotic Bak suggests that sponges may also harbor one or more proteins bearing pro-apoptotic BH3 motif sequences to act in a manner analogous to BH3-only proteins in higher organisms, as indeed has been identified in hydra.24 However, to date no such sponge BH3-only protein has been identified, it may simply be that the BH3 sequence in these proteins lacks sufficient similarity to be recognized, as the BH3 motif is only a short motif with little absolute conservation.7 Alternatively, the BH3-only proteins may have evolved later in evolutionary time. The presence of a Bak-like molecule that can be engaged by a pro-survival BHP2 suggests that apoptosis regulation in sponges is more similar to what is found in mammals compared with, for example, the worm Caenorhabditis elegans, where Bax/Bak orthologs are absent and CED-4 fulfils the role assumed by Apaf-1 in mammals to control caspase activation.25 However, it is noted that C. elegans may be unusual among the worms, with Platyhelminthes26 harboring intrinsic apoptotic machineries that resemble their more modern mammalian relatives.
In summary, our findings suggest that the molecular machinery and mechanisms for executing Bcl-2-mediated apoptosis as observed in mammals are evolutionary ancient, with early regulation of apoptotic machineries closely resembling their modern counterparts in mammals rather than C. elegans or drosophila.
Materials and methods
Cloning, expression and purification
The codon-optimized cDNA of BHP2 (Uniprot accession number Q967D2) encoding for residues 18–200 was synthesized (GenScript, Piscataway, NJ, USA) and cloned into the pGEX-6P1 vector and expressed in E scherichia coli BL21 (DE3) star cells. Cells were grown in 2YT auto induction medium27 containing 1 mg/ml ampicillin at 25 °C in a shaker incubator operating at 160 rev/min for ~24 h. The cells were harvested by centrifugation at 6000 rev/min (JLA 9.1000 rotor, Beckman Coulter Avanti J-E, Beckman Coulter, Mount Waverley, VIC) for 20 min and resuspended in 50 ml lysis buffer A (50 mM Tris pH 8.5, 150 mM NaCl and 1 mM ethylene diamine tetraacetic acid (EDTA)) supplemented with lysozyme. The cells were lysed using one cycle at 35 kPi’s in the benchtop TS cell disrupter (Constant Systems Ltd, Daventry, UK). The lysate was transferred to SS34 tubes for further centrifugation at 16 000 rev/min (JA-25.50 rotor, Beckman Coulter Avanti J-E) for 20 min. The supernatant loaded onto a 2 ml Glutathione Sepharose 4B resin column (GE Healthcare, Aurora, OH, USA) equilibrated with buffer A. After sample application, the column was washed with five column volumes of buffer A followed by HRV 3C protease cleavage on column overnight at 4 °C.
The column was washed with five column volumes of buffer A to remove the liberated target protein and concentrated to a volume of 1 ml. Concentrated BHP2 was subjected to size-exclusion chromatography using a Superdex S200 10/300 column mounted on an ÄKTApure system (GE Healthcare) equilibrated in 25 mM HEPES pH 7.5, 150 mM NaCl, where it eluted as a single peak. The final sample purity was estimated to be higher than 95% based on SDS-PAGE analysis.
Measurement of dissociation constants
Binding affinities were measured using a MicroCal iTC200 system (GE Healthcare) at 25 °C using BHP2 in 25 mM HEPES pH 7.5, 150 mM NaCl at a final concentration of 20 μM. BH3 domain peptide ligands were used at a concentration of 200 μM and titrated using 19 injections of 2.0 μl of ligand. All affinity measurements were performed in triplicate. Protein concentrations were measured using a Nanodrop UV spectrophotometer (Thermo Fisher Scientific, Scoresby, Victoria, Australia) at a wavelength of 280 nm. Peptide concentrations were calculated based on the dry peptide weight after synthesis. The BH3 domain peptides used were commercially synthesized using liquid-phase peptide synthesis (GenScript) and were purified to a final purity of 95%. With the exception of LB-Bak-2 BH3 all peptide sequences were previous described.28 LB-Bak-2 BH3 peptides span residues 62–89 (Uniprot accession code Q1RPT5).
Crystallization and data collection
The complex of BHP2 with LB-Bak-2 BH3 was reconstituted as previously described29 by adding LB-Bak-2 BH3 domain peptide at a 1:1.25 M ratio to BHP2. The reconstituted complex was concentrated to 5 mg/ ml using a 3 kDa molecular weight cut-off centrifugal concentrator (Millipore, Bayswater, Victoria, Australia), flash-cooled and stored under liquid nitrogen. High-throughput sparse matrix screening was carried out using 96-well sitting-drop trays (Swissci, Neuheim, Switzerland) and the vapor diffusion method at 20 °C at the CSIRO Collaborative Crystallization Centre, Melbourne, Australia. The initial crystallization conditions used were from commercially available screening kits (Shotgun Screen, PACT Com7 C3 and JCSG-C3 from C3 CSIRO).
Crystals of BHP2:LB-Bak-2 BH3 were obtained at a protein concentration of 5 mg/ml using the sitting-drop method at 20 °C in 28.7% (w/v) PEG 1500, 0.1 M bis-Tris chloride (Supplementary Figure S1b). The crystals were flash-cooled at −173.15 °C in mother liquor supplemented with 20% (v/v) ethylene glycol. The BHP2:LB-Bak-2 BH3 complex forms clusters of plate-shaped crystals belonging to space group I2 in the monoclinic crystal system.
Diffraction data were collected on the MX2 beamline at the Australian Synchrotron using a ADSC Quantum 315r CCD detector (Area Detector Systems Corporation, Poway, CA, USA) with an oscillation range of 1.0° per frame and a wavelength of 0.9537 Å. Diffraction data were integrated using XDS30 and scaled using AIMLESS.31 Crystals of BHP2:LB-Bak-2 BH3 contained two chains of BHP2 and two chains of LB-Bak-2 BH3 in the asymmetry unit, with a calculated solvent content of 45.73%. The structure was phased by molecular replacement using MrBUMP32 using Bcl-w as a search model (PDB accession code 4k5b33) and an initial model was built using PHENIX Autobuild34 with Resolve and Buccaneer.35 The final BHP2:LB-Bak-2 complex was manually built using Coot36 and refined using PHENIX.37 Details of the data collection and refinement statistics are summarized in Table 1. All images were generated using PyMOL (New York, USA). All software was accessed via SBGrid.38
R.m.s.d. cluster analysis
The structures of Bcl-2 protein family members were selected from the Protein Data Bank for structural comparison, with one representative structure selected for proteins with multiple experimentally determined structures. Structure-based sequence alignment was performed in STRAP.39 The sequence alignment and superimposed coordinates of the different models were loaded into to R program40 using the bio3d package41 for an r.m.s.d. cluster analysis.
Availability of data and material
Coordinate files have been deposited in the Protein Data Bank under the accession codes 5TWA. Raw diffraction images were deposited in the SBGrid Data Bank42 using the accession code doi:10.15785/SBGRID/388.
References
Lasi M, David CN, Bottger A . Apoptosis in pre-Bilaterians: hydra as a model. Apoptosis 2010; 15: 269–278.
Koziol C, Borojevic R, Steffen R, Muller WE . Sponges (Porifera) model systems to study the shift from immortal to senescent somatic cells: the telomerase activity in somatic cells. Mech Ageing Dev 1998; 100: 107–120.
Zmasek CM, Godzik A . Evolution of the animal apoptosis network. Cold Spring Harb Perspect Biol 2013; 5: a008649.
Vaux DL, Haecker G, Strasser A . An evolutionary perspective on apoptosis. Cell 1994; 76: 777–779.
Adams JM, Cory S . The Bcl-2 protein family: arbiters of cell survival. Science 1998; 281: 1322–1326.
Kvansakul M, Hinds MG . Structural biology of the Bcl-2 family and its mimicry by viral proteins. Cell Death Dis 2013; 4: e909.
Kvansakul M, Hinds MG . The structural biology of BH3-only proteins. Methods Enzymol 2014; 544: 49–74.
Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR . The BCL-2 family reunion. Mol Cell 2010; 37: 299–310.
Shamas-Din A, Brahmbhatt H, Leber B, Andrews DW . BH3-only proteins: orchestrators of apoptosis. Biochim Biophys Acta 2011; 1813: 508–520.
Chipuk JE, Green DR . How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol 2008; 18: 157–164.
Czabotar PE, Westphal D, Dewson G, Ma S, Hockings C, Fairlie WD et al. Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell 2013; 152: 519–531.
Green DR, Kroemer G . The pathophysiology of mitochondrial cell death. Science 2004; 305: 626–629.
Luna-Vargas MP, Chipuk JE . The deadly landscape of pro-apoptotic BCL-2 proteins in the outer mitochondrial membrane. FEBS J 2016; 283: 2676–2689.
Wiens M, Krasko A, Muller CI, Muller WE . Molecular evolution of apoptotic pathways: cloning of key domains from sponges (Bcl-2 homology domains and death domains) and their phylogenetic relationships. J Mol Evol 2000; 50: 520–531.
Wiens M, Diehl-Seifert B, Muller WE . Sponge Bcl-2 homologous protein (BHP2-GC) confers distinct stress resistance to human HEK-293 cells. Cell Death Differ 2001; 8: 887–898.
Wiens M, Belikov SI, Kaluzhnaya OV, Schroder HC, Hamer B, Perovic-Ottstadt S et al. Axial (apical-basal) expression of pro-apoptotic and pro-survival genes in the lake Baikal demosponge Lubomirskia baicalensis. DNA Cell Biol 2006; 25: 152–164.
Holm L, Rosenstrom P . Dali server: conservation mapping in 3D. Nucleic Acids Res 2010; 38 (Web Server issue): W545–W549.
Aouacheria A, Rech de Laval V, Combet C, Hardwick JM . Evolution of Bcl-2 homology motifs: homology versus homoplasy. Trends Cell Biol 2013; 23: 103–111.
Vaux DL, Weissman IL, Kim SK . Prevention of programmed cell death in Caenorhabditis elegans by human bcl-2. Science 1992; 258: 1955–1957.
Jabbour AM, Puryer MA, Yu JY, Lithgow T, Riffkin CD, Ashley DM et al. Human Bcl-2 cannot directly inhibit the Caenorhabditis elegans Apaf-1 homologue CED-4, but can interact with EGL-1. J Cell Sci 2006; 119 (Pt 12): 2572–2582.
Lee EF, Chen L, Yang H, Colman PM, Huang DC, Fairlie WD . EGL-1 BH3 mutants reveal the importance of protein levels and target affinity for cell-killing potency. Cell Death Differ 2008; 15: 1609–1618.
Custodio MR, Prokic I, Steffen R, Koziol C, Borojevic R, Brummer F et al. Primmorphs generated from dissociated cells of the sponge Suberites domuncula: a model system for studies of cell proliferation and cell death. Mech Ageing Dev 1998; 105: 45–59.
Domazet-Loso T, Klimovich A, Anokhin B, Anton-Erxleben F, Hamm MJ, Lange C et al. Naturally occurring tumours in the basal metazoan Hydra. Nat Commun 2014; 5: 4222.
Lasi M, Pauly B, Schmidt N, Cikala M, Stiening B, Kasbauer T et al. The molecular cell death machinery in the simple cnidarian Hydra includes an expanded caspase family and pro- and anti-apoptotic Bcl-2 proteins. Cell Res 2010; 20: 812–825.
Horvitz HR . Genetic control of programmed cell death in the nematode Caenorhabditis elegans. Cancer Res 1999; 59 (7 Suppl): 1701s–1706s.
Bender CE, Fitzgerald P, Tait SW, Llambi F, McStay GP, Tupper DO et al. Mitochondrial pathway of apoptosis is ancestral in metazoans. Proc Natl Acad Sci USA 2012; 109: 4904–4909.
Studier FW . Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 2005; 41: 207–234.
Burton DR, Caria S, Marshall B, Barry M, Kvansakul M . Structural basis of Deerpox virus-mediated inhibition of apoptosis. Acta Crystallogr D Biol Crystallogr 2015; 71 (Pt 8): 1593–1603.
Kvansakul M, Czabotar PE . Preparing samples for crystallization of Bcl-2 family complexes. Methods Mol Biol 2016; 1419: 213–229.
Kabsch W . Xds. Acta Crystallogr D Biol Crystallogr 2010; 66 (Pt 2): 125–132.
Evans P . Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 2006; 62 (Pt 1): 72–82.
Keegan RM, Winn MD . MrBUMP: an automated pipeline for molecular replacement. Acta Crystallogr D Biol Crystallogr 2008; 64 (Pt 1): 119–124.
Schilling J, Schoppe J, Sauer E, Pluckthun A . Co-crystallization with conformation-specific designed ankyrin repeat proteins explains the conformational flexibility of BCL-W. J Mol Biol 2014; 426: 2346–2362.
Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr 2008; 64 (Pt 1): 61–69.
Cowtan K . The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr 2006; 62 (Pt 9): 1002–1011.
Emsley P, Lohkamp B, Scott WG, Cowtan K . Features and development of Coot. Acta Crystallogr D Biol Crystallogr 2010; 66 (Pt 4): 486–501.
Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M et al. Towards automated crystallographic structure refinement with phenix refine. Acta Crystallogr D Biol Crystallogr 2012; 68 (Pt 4): 352–367.
Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M et al. Collaboration gets the most out of software. Elife 2013; 2: e01456.
Gille C, Fahling M, Weyand B, Wieland T, Gille A . Alignment-Annotator web server: rendering and annotating sequence alignments. Nucleic Acids Res 2014; 42 (Web Server issue): W3–W6.
R Core TeamRFfSC R: A Language and Environment for Statistical Computing 2016. https://www.r-project.org/.
Grant BJ, Rodrigues AP, ElSawy KM, McCammon JA, Caves LS . Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics 2006; 22: 2695–2696.
Meyer PA, Socias S, Key J, Ransey E, Tjon EC, Buschiazzo A et al. Data publication with the structural biology data grid supports live analysis. Nat Commun 2016; 7: 10882.
Liu X, Dai S, Zhu Y, Marrack P, Kappler JW . The structure of a Bcl-xL/Bim fragment complex: implications for Bim function. Immunity 2003; 19: 341–352.
Yan N, Gu L, Kokel D, Chai J, Li W, Han A et al. Structural, biochemical, and functional analyses of CED-9 recognition by the proapoptotic proteins EGL-1 and CED-4. Mol Cell 2004; 15: 999–1006.
Campbell S, Thibault J, Mehta N, Colman PM, Barry M, Kvansakul M . Structural insight into BH3 domain binding of vaccinia virus antiapoptotic F1L. J Virol 2014; 88: 8667–8677.
Acknowledgements
We thank MX2 beamline staff at the Australian Synchrotron for help with X-ray data collection, and Janet Newman and Shane Seabrook at the CSIRO C3 Collaborative Crystallization Centre for assistance with crystallization and Cody Hall for technical assistance. This work was supported by the Australian Research Council (Fellowship FT130101349 to MK).
Author contributions
Acquisition of data, analysis and interpretation of data, drafting and revising the article: Sofia Caria; Conception and design, analysis and interpretation of data, drafting and revising the article: Mark Hinds; Conception and design, acquisition of data, analysis and interpretation of data, drafting and revising the article: Marc Kvansakul.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Oberst
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Caria, S., Hinds, M. & Kvansakul, M. Structural insight into an evolutionarily ancient programmed cell death regulator – the crystal structure of marine sponge BHP2 bound to LB-Bak-2. Cell Death Dis 8, e2543 (2018). https://doi.org/10.1038/cddis.2016.469
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2016.469
This article is cited by
-
Diversity in the intrinsic apoptosis pathway of nematodes
Communications Biology (2020)
-
A structural investigation of NRZ mediated apoptosis regulation in zebrafish
Cell Death & Disease (2018)