Abstract
Unconventional prefoldin RPB5 interactor (URI), which acts as an oncoprotein in solid tumors, is associated with RNA polymerase II subunit 5. However, its impact on multiple myeloma (MM) has not been determined. We demonstrate here that URI is overexpressed in MM compared with plasma cells derived from healthy volunteers. Side population (SP) cells sorted from MM cells showed a much higher level of URI than non-SP cells. Using lentivirus-delivered shRNA, we established stable URI knockdown MM cell lines. URI inhibition significantly attenuated the proliferation of MM cells and decreased colony formation compared with the control cells. Tumor growth assays in NOD/SCID mice further confirmed the promotion role of URI during MM development in vivo. Furthermore, URI knockdown markedly reduced the abundance of SP in MM cell lines and enhanced the chemotherapeutic sensitivity of MM towards bortezomib. Mechanically, URI appears to be critically involved in modulating STAT3 activity through regulating interleukin (IL)-6 transcription via interaction with NFκBp65. In conclusion, URI may have an important role in the development of MM and chemotherapeutic resistance through activating the IL-6/STAT3 pathway.
Similar content being viewed by others
Main
Multiple myeloma (MM) is a hematologic malignancy characterized by the clonal expansion of plasma cells in the bone marrow, which produce monoclonal immunoglobulins and lead to complications such as anemia, bone destruction, hypercalcemia, and renal failure.1, 2 MM cases account for ∼10% of all hematologic cancers and ∼2% of all cancer deaths.3 Despite novel insights into the pathobiology of the disease and the availability of new research platforms and therapeutics,4, 5, 6 nearly all patients will eventually relapse or become refractory to drug treatment. Thus, innovative treatment strategies are urgently needed. A major area of investigation is the human bone marrow microenvironment, which has an essential role in promoting growth, survival, and drug resistance in MM.7, 8, 9
The growth and survival of MM cells is highly dependent upon the presence of certain growth-promoting cytokines in the bone marrow microenvironment. One of the most important MM survival factors is the pleiotropic cytokine interleukin-6 (IL-6). IL-6 was originally described as a factor that induces the differentiation of normal B cells to antibody-secreting plasma cells and promotes MM cell growth.10, 11, 12 Whereas the main producers of IL-6 in the bone marrow microenvironment are most likely the bone marrow stromal cells, some MM cell lines both produce and respond to IL-6, supporting an autocrine proliferation loop.10, 13, 14, 15 Importantly, autocrine IL-6 production was shown to be associated with a highly malignant phenotype, a high proliferative index, and resistance to drug-induced apoptosis.15 In addition, serum IL-6 and C-reactive protein, an acute phase protein synthesized in the liver in response to IL-6, are prognostic factors in MM and provide the basis for a biologically based staging method for MM.16, 17 Strategies to target the effects of IL-6 in MM in the clinic have included the use of monoclonal anti-IL-6 alone or in combination with glucocorticoids.18, 19 However, the molecular basis underlying active IL-6 expression in primary MM samples and MM cell lines has not been elucidated.
Unconventional prefoldin RPB5 interactor (URI) is associated with RNA polymerase II subunit RPB5. The URI gene was first isolated and cloned from a human HepG2 cDNA library in 1998 and counteracted transactivation by hepatitis B Virus X Protein.20 URI is also required to maintain genome stability through its important function in regulating the cell cycle.21 Depletion of URI increased cisplatin- and rapamycin-induced apoptosis in ovarian cancer cells by activating mitochondrial S6K1-BAD signaling,22 and overexpression of URI had an anti-apoptotic effect in the proliferation and growth of HCC cells.23 However, the role of URI in the regulation of MM remains to be determined. In this study, we determined the effect of URI in MM on cell proliferation, chemotherapy resistance, survival in vitro, and growth in vivo.
Results
URI overexpresses in MM and MM cell lines
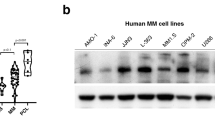
We investigated the expression levels of URI in MM and normal plasma cells separated from bone marrow. The mRNA levels were significantly higher in MM than in normal plasma cells (Figure 1a). URI mRNA was constitutively expressed at higher levels in most MM cell lines than normal plasma cells (Figure 1b). We examined the URI protein content in several MM cell lines and found an expression pattern similar to mRNA expression (Figure 1c). URI protein expression was also observed in MM cells in bone marrow specimens of patients with MM (Figure 1d). Interestingly, URI protein levels were upregulated substantially in MM cells from almost all patients with MM (20/22), whereas the normal plasma cells contained a very low level of URI (Figure 1e). These results demonstrate that URI is overexpressed in MM cells from MM patients compared with normal plasma cells.
URI is overexpressed in MM and MM cell lines. (a) URI mRNA expression in normal plasma cells from five volunteers (Nor) and MM cells from 12 MM patients or (b) five MM cell lines. CD138+ cells enriched from bone marrow of healthy volunteers by immunomagnetic cell sorting method (MACS). URI mRNA in the cells was determined by quantitative real-time PCR. (c) Western blotting analysis of the expression of the URI protein in MM cell lines. The URI protein levels were quantified relative to the loading control. (d) Representative immunohistochemistry staining of URI in the bone marrow of volunteers and patients with MM. (e) The percentage of high expression of URI in normal bone marrow and patients with MM. Error bars represent the S.E.M. *P<0.05
Knocked down URI inhibits MM cell growth in vitro and in vivo
To observe the function of URI in MM, the expression of URI was knocked down in MM cell lines. As shown in Figures 2a and b, lentivirus-delivered shRNA significantly inhibited the mRNA and protein levels of URI in NCI-H929 and LP-1 cells. In the stably transfected cell lines, the viable cell numbers were determined by the CCK-8 assay (Figure 2c). Suppression of URI resulted in a significant inhibition of MM cell line growth in normal culture conditions. Interestingly, growth inhibition was associated with lower numbers of URI-shRNA cells in the G2/M stage than in the control cells (Figure 2d). Next, we determined whether depletion of URI interfered with the clonogenic growth and survival of MM cells. As shown in Figure 2e, shRNA-URI MM cells suppressed colony formation compared with control cells. The effect of URI on the tumorigenic potential of MM cells was evaluated in vivo. NCI-H929 and LP-1 cells were transduced with a lentivirus expressing an shRNA targeting URI or the empty vector. These cells were inoculated in NOD/SCID mice, and tumor formation was monitored (Figure 2f). As shown in Figure 2g, knocked down URI markedly reduced the growth of MM xenografts compared with the control group in vivo.
Knockdown of URI inhibits MM cell growth in vitro and in vivo. (a and b) URI expression was analyzed by real-time PCR and immunoblotting in URI knockdown (shURI) and control cells (NC). The URI protein levels were quantified relative to the loading control. (c) Cell proliferation was detected in control (NC) and shURI MM cells.The MM cells (2 × 103 cells per well) were seeded in 96-well plates and cultured overnight at 37 °C. Cell proliferation was detected by a CCK-8 assay at various time points according to the manufacturer’s instructions. (d) Flow cytometric analysis of the proportion of cells in G2/M phase in shURI MM and control cells (NC). Representative results are shown. (e) Quantification of methylcellulose colony formation of NCI-H929 and LP-1 after URI knockdown (shURI) of three independent experiments. Myeloma cells were adjusted to a concentration of 1 × 103 per ml cells using human methylcellulose-based medium according to the manufacturer’s instructions and were then plated in 35-mm2 culture dishes. Colonies were counted by microscopy 10 days after plating. (f and g) The tumors excised from NOD/SCID mice after being inoculated subcutaneously with shURI MM cells and their control cells (NC) for 15 days. Tumor size was measured once every 3 days, and tumor volume was calculated by the formula: (width)2 × length/2. Error bars represent the S.E.M. *P<0.05
URI expression maintains the frequency of side population (SP) cells in MM
SP cells exhibit key characteristics of cancer-initiating cells, including capacities for differentiation, repopulation, clonogenicity, and self-renewal. We screened for SP cells in human MM cell lines, based on their ability to export Hoechst 33342 dye. Flow cytometry revealed SP cells in the tail region of the dot plot. Figure 3a depicts SP cells from the RPMI-8226, NCI-H929, and LP-1 lines: low-level accumulation of intracellular Hoechst 33342 is characteristic of SP cells and the population disappears when the cells are treated with the ATP-binding cassette (ABC) transporter inhibitor verapamil. URI expression levels were significantly increased in the SP cells compared with the non-SP cells (Figure 3b). The expression pattern of URI indicates that it may influence the frequency of SP in MM. We next analyzed the SP size in URI-shRNA and control cells in NCI-H929 and LP-1 and found that knockdown of URI markedly reduced the percentage of SP in both MM cell lines (Figure 3c). Furthermore, URI inhibition led to decreased levels of ABCG2, an ABC membrane transporter that gives these cells a distinct staining pattern with Hoechst 33342 (Figure 3d). MM SP cells exhibit cancer stem cell-like characteristics, and they have greater tumorigenic potential than non-SP cells. We then compared the expression of stemness-associated genes between URI knockdown cells and control cells (Figures 3e and f). Consistently, the downregulation of stem cell-associated genes, including KLF4, SOX2, OCT4, C-MYC, BMI1, and NOTCH1, was observed in URI knockdown cells in LP-1 and NCI-H929 cells.
URI maintains the frequency of SP cells in MM. (a) Flow cytometric analysis of the frequency of SP cells in NCI-H929, LP-1, and RPMI-8226 stained with Hoechst 33342. (b) The URI mRNA levels in SP and non-SP cells were detected by real-time RT-PCR. (c) Flow cytometric analysis of the frequency of SP cells in NCI-H929 or LP-1 cells with URI knockdown (shURI) and control cells (NC). (d) The mRNA expression of ABCG2 in URI knockdown (shURI) and control cells (NC) by real-time RT-PCR. (e and f) The mRNA expression of five genes related to stemness in LP-1 or NCI-H929 shURI and control cells (NC) by real-time RT-PCR. The data are mean±S.E.M. and are representative of three independent experiments. *P<0.05; **P<0.01; ***P<0.001
Inhibition of URI enhances bortezomib-induced MM cell death
Next, we investigated the influence of URI on the chemotherapeutic sensitivity of MM treatment. Bortezomib, a proteasome inhibitor, has significant anti-myeloma activity both in newly diagnosed and refractory/relapsed MM.24, 25 Bortezomib treatment induced cell growth arrest and cell death. Blocking the expression of URI significantly enhanced the therapeutic effects of bortezomib both in LP-1 and NCI-H929 cells (Figure 4a). Further, as detected by Annexin V staining, apoptosis induced by bortezomib was observed in the two cell lines (Figures 4b and c). As the results show, knockdown of URI markedly increased apoptotic cell number in LP-1 and NCI-H929 cells upon bortezomib treatment. To determine the degree to which bortezomib reduced SP cell number, we assessed the %SP after exposing cells to bortezomib (Figure 4d). We found that bortezomib markedly reduced the %SP among both LP-1 and NCI-H929 cells and that URI-shRNA cell contained a lower SP than control cells. Western blotting assays revealed more cleaved PARP in URI-shRNA cells than in control cells upon bortezomib treatment (Figures 4e and f). These results suggest that URI regulates chemotherapy sensitivity of MM towards bortezomib via maintaining the frequency of SP cells.
Inhibition of URI enhances bortezomib-induced MM cell death. (a) Cell proliferation was detected by a CCK-8 assay at various bortezomib concentrations for 24 h. (b and c) LP-1 NC/shURI and NCI-H929 NC/shURI cells were incubated in the presence of 10 nM bortezomib for 24 h and apoptotic cells were determined by AnnexinV-FITC and propidium iodide staining. (d) Flow cytometric analysis of the frequency of SP cells in URI knockdown (shURI) and control cells (NC) after being exposed to 20 nM bortezomib for 24 h. (e and f) Immunoblotting for PARP in LP-1 NC/shURI and NCI-H929 NC/shURI cells after being treated with different concentrations of bortezomib for 24 h. The clevated PARP protein levels were quantified relative to the loading control
Suppression of URI attenuates IL-6 expression level and STAT3 activity
Of the various secreted cytokines, paracrine and autocrine regulation by IL-6 has a particularly important role in myeloma cell proliferation and chemoresistance. We examined IL-6 expression in URI wild-type and knockdown MM cells. Luciferase reporter assays indicated that the downregulation of IL-6 by knocked down URI was achieved through inhibiting the activity of the IL-6 promoter activity in LP-1 and NCI-H929 cells (Figure 5a). Quantitative real-time PCR assays also demonstrated that the inhibition of URI attenuated IL-6 transcription (Figure 5b). Next, we used ELISA to detect IL-6 concentration in the medium supernatant (Figure 5c) and found that suppression of URI significantly decreased the IL-6 protein level in the cell culture medium.
Suppression of URI attenuates IL-6 expression and STAT3 activity. (a) Knockdown of URI in LP-1 and NCI-H929 cells inhibited the activity of the IL-6 promoter by the luciferase reporter assay. MM cells were transfected with the IL-6 reporter plasmids (0.2 μg), together with the control plasmid pRL-TK (0.02 μg). After 24 h, cells were lysated and then luciferase activities were detected. (b) Knockdown of URI in LP-1 and NCI-H929 cells resulted in downregulation of IL-6. Total RNA was extracted from the control and URI knockdown cells, and IL-6 mRNA was determined by real-time PCR. (c) Knockdown of URI in LP-1 and NCI-H929 cells resulted in downregulation of IL-6 concentration in the culture supernatant at indicated times as assayed using ELISA. (d) The IL-6 mRNA expression in SP cells and non-SP cells by real-time RT-PCR. (e) Knockdown of URI inhibited IL-6 (20 ng/ml) induced phosphorylation of STAT3 in LP-1 and NCI-H929 cells. The p-STAT3 protein levels were quantified relative to the total protein levels of STAT3. (f) Luciferase reporter assay of NF-κB was assessed in LP-1 NC/shURI and NCI-H929 NC/shURI cells. Error bars represent the S.E.M. from three independent experiments. (g) Knockdown of URI inhibited TNFα (10 ng/ml)-induced phosphorylation of p65 in LP-1 and NCI-H929 cells. The p-p65 protein levels were quantified relative to the total protein levels of p65. (h) ChIP assays indicated a significant decrease in p65 bound upstream of the transcriptional start site of IL-6 upon URI knockdown (shURI) in LP-1 or NCI-H929 cells compared with corresponding vector control cells (NC). Normal rabbit IgG served as a negative control. (i) NCI-H929 cells were co-transfected with Flag-URI and Flag-p65. Forty-eight hours after transfection, the cells were treated with TNFα at a concentration of 10 ng/ml for 20 min before harvesting. Whole cell lysates were immunoprecipitated with anti-URI, anti-p65 antibody, or mouse IgG (negative control) and then were analyzed by western blotting. The cell lysates were also subjected to immunoblotting (lower panel). Error bars represent the S.E.M. from at least three independent experiments. *P<0.05; **P<0.01; ***P<0.001
Our results suggest that URI influences SP size in MM; therefore, we examined IL-6 mRNA levels in SP and non-SP cells (Figure 5d). The data indicated that SP cells in the LP-1 and NCI-H929 cell lines expressed IL-6 to much higher levels than non-SP cells. We treated MM cells with IL-6 and found that suppression of URI significantly attenuated phosphorylation of STAT3 (Figure 5e), indicating the role of URI in the regulation of IL-6/STAT3 signaling activation.
URI upregulates IL-6 expression through an interaction with NFκB p65
IL-6 is one of the most important NFκB-dependent cytokines. To assess the effect of URI on NFκB, we measured NFκB activity by a luciferase reporter assay (Figure 5f). The results indicated that URI knockdown significantly attenuated the activity of NFκB in MM cells. A western blotting assay also demonstrated that URI-shRNA LP-1 and NCI-H929 cells showed a much lower level of phosphorylation of NFκB p65 than control cells stimulated by TNFα (Figure 5g), indicating that the inhibition of URI could decrease NFκB activity. To further examine the different potential binding between p65 and the IL-6 promoters in control and URI-shRNA cells, a ChIP-qPCR assay was performed (Figure 5h). After URI knockdown, a decrease in p65 binding to IL-6 promoters in LP-1 and NCI-H929 cells was observed. The co-immunoprecipitation assay revealed that URI could associate with p65, which is a key component of the transcriptional preinitiation complex (PIC) of IL-6, and the interaction was markedly increased when the cells were treated with TNFα (Figure 5i). In addition, we undertook a co-immunoprecipitation assay between URI and STAT3 or CREB, which are important transcription factors for IL-6 expression.26, 27, 28 No interactions were observed between URI and the two transcription factors (data not shown). These results suggest that URI regulates IL-6 expression through enhancing NFκB p65 transcriptional efficiency.
Discussion
An important challenge in MM research lies in pinpointing the critical factors that regulate growth and survival so that potential therapeutic targets can be defined. One of these factors is the pleiotropic cytokine IL-6, a cytokine mediating diverse biological functions in normal tissues and of particular importance in the promotion of myeloma cell expansion and survival.10, 29 Although research suggests that the main producers of IL-6 in the bone marrow microenvironment are most likely the bone marrow stromal cells, many MM cells both produce and respond to IL-6, supporting an autocrine mechanism sustaining high levels of proliferation of MM cells.13, 14, 15 IL-6 activates multiple cell survival and proliferation signaling pathways in myeloma cells, including the Janus kinase/signal transducer and activator of transcription 3 (JAK/STAT3) pathway,30, 31 the mitogen-activated protein kinase kinase/extracellular signal-related kinase (MEK/ERK) pathway, and the phosphoinositide 3-kinase (PI3-K)/Akt pathway.32, 33 In addition, autocrine IL-6 production was shown to be associated with a resistance to chemical agent-induced cell death.15, 34 Interestingly, the present study revealed that URI was a functional regulator of IL-6 transcription in MM. It has been widely reported that STAT3, CREB, and p65 are the major transcription factors for IL-6.26, 27, 28 However, we did not find an interaction between URI and STAT3 or CREB. Nevertheless, URI was observed in association with NFκB p65, which is assembled in the PIC of IL-6.
Our results also indicated that URI could regulate NFκB activation in MM cells. The NFκB transcription factor has been at the center of studies investigating MM pathophysiology.35 Constitutively active NFκB has been observed in primary MM samples, and the IκB kinase inhibitor, which blocks TNFα-induced NFκB activation, was shown to decrease MM cell growth and survival in vitro.36, 37 The implication of NFκB in MM is supported by the observation that the proteasome inhibitor bortezomib, which blocks degradation of the NFκB inhibitor protein IκB, has been successfully used in the treatment of newly diagnosed and refractory/relapsed MM.38 An intriguing finding of the present work was that URI regulated the anti-MM effects of bortezomib: in fact, inhibition of URI increased bortezomib-induced cytotoxicity in MM cells, whereas the presence of URI dampened bortezomib-induced apoptosis. Meanwhile, our data indicated that URI modulated the transcriptional function of NFκB p65, which may contribute to the attenuation of bortezomib-induced cytotoxicity on MM. These results underscore the relevance of URI to the induction of apoptosis by bortezomib. Our results may therefore contribute to the understanding of the intracellular functions of URI and of the biochemical perturbations underlying the apoptotic stimuli induced by URI, complementing previous observations on the anticancer activity of this protein.
Recent studies have revealed the presence of SP cells in many types of cancer, including MM.39, 40 Here, we found that URI expression levels were significantly increased in SP cells compared with non-SP cells and that knockdown of URI markedly reduced the abundance of SP in MM cell lines. These results strongly indicate that the presence of URI maintains the frequency of SP cells in MM. SP cells in various types of cancer showed significantly higher potential to initiate tumors in NOD/SCID mice than their non-SP counterparts. They are also more likely to be resistant to certain anticancer drugs than non-SP cells. In the present study, URI was found to enhance MM cell growth in vitro and in vivo and to regulate the chemotherapeutic sensitivity of MM towards bortezomib treatment. The regulatory role of URI on SP may partly contribute to its biological function in MM. Accumulating evidence suggests that SP cells, a small population of cells from cancer cell lines, are enriched in a subset of cancer stem-like cells. They are responsible for tumor initiation, metastasis, and recurrence.40, 41, 42 Although the mechanism for producing the SP phenotype is unclear, it is believed that efflux of the DNA-binding dye Hoechst 33342 by the ABCG2/BCRP transporter is required for detection of the ‘side population’ phenotype that is characteristic of stem cells from many tissues.43 Our data showing that inhibition of URI strongly reduced the downregulation of ABCG2 expression in MM cell lines also confirmed the regulatory function of URI on SP cells.
STAT3 has been identified as a pro-survival protein activated downstream of the MM growth and survival factor IL-6 and as a promising drug target. We used IL-6 to treat MM cells and found that knockdown of URI significantly attenuated the phosphorylation of STAT3, indicating that URI could increase IL-6-induced STAT3 activation. However, the exact mechanism of URI on STAT3 needs additional exploration. An important issue influencing the activation is the response of MM cells to IL-6 stimulation. Previous reports have classified MM cells into IL-6 dependent and -independent groups according to whether the cells are dependent on IL-6 for survival or proliferation. There are some cell lines that are independent of IL-6 both for survival and proliferation. After treating several MM cell lines with IL-6, we detected the level of phosphorylated STAT3 and found that most MM cell lines responded to IL-6, even though some were IL-6-independent cells. Therefore, modulating IL-6/STAT3 signaling is a potential therapeutic strategy for myeloma not only characterized by the pathological IL-6-dependent type but also by IL-6-responsive MM.
Taken together, our observations highlight a novel exciting scenario and provide new formal evidence for a major role of URI on MM cell proliferation, survival, and chemotherapeutic resistance. The data presented here identified URI as a regulator of IL-6 transcription. As the significant role of IL-6 is inducing transformation and development of MM, targeting URI in myeloma may dramatically lower IL-6 production, which is an attractive therapeutic strategy against this disease.
Materials and Methods
Reagents
The following reagents were purchased from the indicated manufacturers: rh IL-6 and TNFα were obtained from PEPROTECH (Rocky Hill, NJ, USA) and anti-P-STAT3, anti-STAT3, anti- NFκB p-p65, anti-NFκBp65, and anti-PARP antibodies were obtained from Cell Signaling Technology (Boston, MA, USA). Anti-GAPDH antibody was purchased from Santa Cruz Biotechnology (Dallas, TX, USA) and the anti-URI antibody was from Proteintech (Chicago, IL, USA). Bortezomib was purchased from Selleckchem (Houston, TX, USA).
Cells and cultures
Human MM cell lines NCI-H929, LP-1, RPMI-8226, and U266 were used in this study. NCI-H929 was obtained from Dr. Margaret HL Ng (Prince of Wales Hospital, Chinese University of Hong Kong). LP-1-secreting IgG l light chain was kindly provided by Dr. Hallek (Laboratorium für Molekulare Biologie, Genzentrum, Ludwig-Maximilians-Universität München, Germany). RPMI-8226 (CCL-155) and U266 (TIB-196) were obtained from American Type Culture Collection (ATCC). The cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mM of L-glutamine (Sigma, St. Louis, MO, USA), 100 U/ml of penicillin G, and 100 mg/ml of streptomycin (Sigma). For depletion of URI, plasmids containing shURI and lentivirus-shURI and control virus were purchased from Genechem Co. (Shanghai, China). MM cells were seeded into wells. After incubating for 24 h, these cells were transfected with plasmid or lentivirus according to the manufacturer’s protocol.
Patient samples
Primary myeloma cells were isolated from the bone marrow samples of 12 MM patients receiving routine diagnostic aspiration, and normal plasma cells were isolated from five healthy volunteers with informed consent approved by the Institutional Ethics Committee. Briefly, cells were separated by Ficoll density gradient centrifugation and washed in phosphate-buffered saline (PBS) twice before incubation with an anti-CD138 antibody coupled to magnetic beads (Miltenyi Biotechm, Aurburn, CA, USA). The cells were selected using a magnetic affinity column according to the manufacturer’s recommendation. The percentage of CD138+ cells isolated from bone marrow of normal donors was 0.5∼2% in mononuclear cells. The purity of the cell preparation was verified to be 95% by fluorescence-activated cell-sorting (FACS) analysis and light microscopy. Bone marrow tissues were obtained from 22 MM patients who received spinal surgery because of the disease and five patients who received spinal surgery because of spinal trauma, with informed consent approved by the institutional ethics committee. Briefly, bone marrow samples were fixed in formalin for 24 h, decalcified, embedded in paraffin, and sliced. All procedures involving human specimens were undertaken after written informed consent according to the Declaration of Helsinki and using a protocol approved by the Institutional Review Board for human protection.
Immunohistochemistry
Immunohistochemistry was performed as described previously.44 The tissue sections were dewaxed, rehydrated and then immersed in methanol containing 0.3% hydrogen peroxide (Sinopharm Chemical Reagent Co., Ltd.) for 30 min to block endogenous peroxidase activity. Subsequently, the sections were heated in a pressure cooker filled with 10 mM ethylenediaminetetraacetic (EDTA Sigma) buffer (pH 8.0) for 2 min. After cooling, the sections were incubated in 1% blocking serum (Chemicon) for 30 min to reduce nonspecific binding. The sections were incubated in the primary polyclonal antibodies against URI and IL-6. The visualization signal was developed with diaminobenzidine and the slides were counterstained in hematoxylin. Stained sections were evaluated in a blinded manner without prior knowledge of the clinical information using the German immunoreactive score (IRS) as described previously.45 Briefly, the IRS assigns sub-scores for immunoreactive distribution (0–4) and intensity (0–3), then multiplies them to yield the IRS score. The percent positivity was scored as ‘0’ (<5%), ‘1’(5-25%), ‘2’ (25-50%), ‘3’ (50-75%) or ‘4’ (>75%). The staining intensity was score as ‘0’ (no staining), ‘1’ (weakly stained), ‘2’ (moderately stained) or ‘3’ (strongly stained). The final URI expression score was calculated with the value of percent positivity score plus staining intensity score, which ranged from 0 to 12. We estimated IRS by averaging the values in eight fields at × 400 magnification for each specimen. URI expression was defined as follows: low expression (score 0–6/0–3) and high expression (score >6/4–6). Cases with discrepancies in IRS score were discussed with other pathologists until a consensus was reached.
Western blot and co-immunoprecipitation assays
Whole cell extracts were prepared in RIPA buffer and centrifuged at 12 000 g for 15 min. Protein concentrations were measured using the bicinchoninic acid assay. Immunoblotting was performed using specific primary antibodies, and immune-complexes were incubated with the fluorescein-conjugated secondary antibody and then detected using an Odyssey fluorescence scanner (Li-Cor, Lincoln, NE). For co-immunoprecipitation experiments, cell lysates were prepared in RIPA buffer and protein concentrations were measured. The lysates were incubated with 2 μg anti-URI, or anti-p65 and normal mouse immunoglobulin G (Santa Cruz Biotechnology) for 8 h at 4 °C, followed by the addition of Protein A/G Plus-Agarose (Santa Cruz Biotechnology) for another 4 h. The beads were centrifuged and washed 5 times with lysis buffer and resuspended in equal volume of RIPA buffer. The samples from these assays were analyzed by western blotting.
RNA collection, cDNA synthesis, and real-time PCR analysis
Total RNA was extracted from fresh-frozen primary myeloma cells, normal plasma cells, and cell lines in Trizol (Invitrogen, Carlsbad, CA, USA). Reverse transcription of total RNA was performed using random hexamers (Roche Diagnostics, Penzberg, Germany) and SuperScriptII reverse transcriptase (Invitrogen). Polymerase chain reaction (PCR) amplifications of the respective genes were carried out with 40 ng complementary DNA, 500 nM forward and reverse primers, and iTaqSYBRGreen Supermix (Bio-Rad Laboratories, Hercules, CA, USA) in a final volume of 10 μl. The sequences of the PCR primers used in this study are shown in Table 1.
ELISA assay
ELISA assays were performed using the conditioned medium collected from 2-day cultures of cells seeded at 2 × 105 cells/plate, and then analyzed by a human IL-6 kit purchased from Dakewei Biotech Company (Shanghai, China).
Cell viability assays
Cell viability was determined by the Cell Counting Kit-8 assay (DOJINDO, Kumamoto, Japan) according to the manufacturer’s instructions. Briefly, 5 × 103 cells were incubated in 96-well plates with or without treatments in culture medium for different time points, and then 10 μl of the CCK-8 solution was added to each well. After a 1-h incubation at 37 °C, the optical density (OD) was measured using a spectrophotometer (Molecular Devices Co., Sunnyvale, CA, USA) and the fold-increase in the OD compared with that of the control (proliferation index) was calculated. All experiments were performed in triplicate. The absorbance of each well was measured at 450 nm with a microtiter plate reader. Apoptosis was evaluated by staining the cells with an AnnexinV-FITC and propidium iodide labeling kit (Invitrogen) according to the manufacturer’s instructions.
Colony formation assays
Briefly, myeloma cells were adjusted to a concentration of 1 × 103 per ml cells using human methylcellulose-based medium (R&D Systems Inc.) according to the manufacturer’s instructions and were then plated in 35-mm2 culture dishes. The cells were incubated at 37 °C and 5% CO2. Colonies consisting of more than 40 cells were counted using microscopy 10 days after planting.
Chromatin immunoprecipitation (ChIP) assay
Cells (5 × 106) were fixed in 0.5% formaldehyde for 10 min and lysed. Chromatin was sonicated to an average length of 0.2–0.5 kb. Chromatin was then immunoprecipitated with anti-p65 antibody, which recognizes transcriptionally active chromatin regions, or beads-only for the negative control. After washing, crosslinking was reversed and DNA was purified with phenol/chloroform and ethanol precipitation. Isolated DNA was analyzed using qRT-PCR. The following primers were used for amplification of the IL-6 promoter and β-actin control region: IL-6-ChIP-fw: 5-CTAGTTGTGTCTTGCCATGC-3, IL-6-ChIP-rv: 5-CAGAATGAGCCTCAGACATC3, β-actin-ChIP-fw: 5-TCCACCTTCCAGCAGATGTG-3, and β-actin-ChIP-rv: 5-GCAACTAAGTCATAGTCCGCCTAGA-3.
Selection and characterization of SP cells
To identify the SP within the population of MM cells, the cells were adjusted to a final cell density of 1 × 106 cells/ml in RPMI 1640 medium containing 2% FBS and 10 mM HEPES buffer (Invitrogen). Hoechst 33342 (Sigma, 1 mg/ml) was then added to a final concentration of 5 μg/ml followed by incubation in a water bath at 37 °C for 90 min with intermittent shaking. As a negative control, the sample was preincubated with 50 μmol/l verapamil. At the end of incubation, the cells were incubated on ice for 10 min, after which they were washed and resuspended in ice-cold PBS (Dulbecco’s PBS without calcium and magnesium) with 2% FBS in the dark. To gate only viable cells, a propidium iodide solution (2 μg/ml) was added to the cells before analysis. The cells were then filtered through a 70-μm filter to obtain a single cell suspension, which was analyzed and sorted on a Moflo cell sorter (Beckman Coulter, Brea, CA, USA). The Hoechst 33342 dye was excited at 357 nm and its fluorescent emission was measured at 402–446 and 650–670 nm. All experiments were performed at least three times.
Xenograft studies
Six- to 8-week-old male NOD/SCID mice were injected subcutaneously in the bilateral flanks with 2 × 107 myeloma cells in 100 μl PBS stably transduced with control lentivirus (right flanks, n=6) and URI shRNA lentivirus (left flanks, n=6). Tumor size was measured once every 3 days using a caliper, and tumor volume was calculated by the formula: (width)2 × length/2. After 15 days, the mice were euthanized and the tumors were excised.
Statistical analysis
The data shown represent the mean values of at least three independent experiments and are expressed as the mean±S.D. Statistical analyses were performed by Student’s t-test, using the statistical software GraphPad Prism 4 (GraphPad Software Inc., La Jolla, CA, USA). Statistical significance was set at a level of P<0.05. SPSS 15.0 software (SPSS Inc., Chicago, IL, USA) was also used for statistical analyses and a P-value <0.05 was considered significant.
Abbreviations
- ATCC:
-
American Type Culture Collection
- ChIP:
-
chromatin immunoprecipitation
- IL-6:
-
interleukin-6
- IRS:
-
German immunoreactive score
- MM:
-
multiple myeloma
- SP:
-
side population
- PI3-K:
-
phosphoinositide 3-kinase
- PIC:
-
transcriptional preinitiation complex
- STAT3:
-
signal transducer and activator of transcription 3
- URI:
-
unconventional prefoldin RPB5 interactor
References
Anderson KC, Alsina M, Bensinger W, Biermann JS, Chanan-Khan A, Cohen AD et al. Multiple myeloma. J Natl Compr Canc Netw 2011; 9: 1146–1183.
Podar K, Richardson PG, Hideshima T, Chauhan D, Anderson KC . The malignant clone and the bone-marrow environment. Best Pract Res Clin Haematol 2007; 20: 597–612.
Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A et al. Cancer statistics 2005 CA Cancer J Clin 2005; 55: 10–30.
Fonseca R, Stewart AK . Targeted therapeutics for multiple myeloma: the arrival of a risk-stratified approach. Mol Cancer Ther 2007; 6: 802–810.
San-Miguel J, Harousseau JL, Joshua D, Anderson KC . Individualizing treatment of patients with myeloma in the era of novel agents. J Clin Oncol 2008; 26: 2761–2766.
Ocio EM, Mateos MV, Maiso P, Pandiella A, San-Miguel JF . New drugs in multiple myeloma: mechanisms of action and phase I/II clinical findings. Lancet Oncol 2008; 9: 1157–1165.
Podar K, Chauhan D, Anderson KC . Bone marrow microenvironment and the identification of new targets for myeloma therapy. Leukemia 2009; 23: 10–24.
Novak AJ, Darce JR, Arendt BK, Harder B, Henderson K, Kindsvogel W et al. Expression of BCMA, TACI, and BAFF-R in multiple myeloma: a mechanism for growth and survival. Blood 2004; 103: 689–694.
Moreaux J, Legouffe E, Jourdan E, Quittet P, Reme T, Lugagne C et al. BAFF and APRIL protect myeloma cells from apoptosis induced by interleukin 6 deprivation and dexamethasone. Blood 2004; 103: 3148–3157.
Kawano M, Hirano T, Matsuda T, Taga T, Horii Y, Iwato K et al. Autocrine generation and requirement of BSF-2/IL-6 for human multiple myelomas. Nature 1988; 332: 83–85.
Hirano T . Interleukin 6 and its receptor: ten years later. Int Rev Immunol 1998; 16: 249–284.
Ishikawa H, Tsuyama N, Abroun S, Liu S, Li FJ, Taniguchi O et al. Requirements of src family kinase activity associated with CD45 for myeloma cell proliferation by interleukin-6. Blood 2002; 99: 2172–2178.
Urashima M, Ogata A, Chauhan D, Vidriales MB, Teoh G, Hoshi Y et al. Interleukin-6 promotes multiple myeloma cell growth via phosphorylation of retinoblastoma protein. Blood 1996; 88: 2219–2227.
Chauhan D, Uchiyama H, Akbarali Y, Urashima M, Yamamoto K, Libermann TA et al. Multiple myeloma cell adhesion-induced interleukin-6 expression in bone marrow stromal cells involves activation of NF-kappa B. Blood 1996; 87: 1104–1112.
Frassanito MA, Cusmai A, Iodice G, Dammacco F . Autocrine interleukin-6 production and highly malignant multiple myeloma: relation with resistance to drug-induced apoptosis. Blood 2001; 97: 483–489.
Bataille R, Jourdan M, Zhang XG, Klein B . Serum levels of interleukin 6, a potent myeloma cell growth factor, as a reflect of disease severity in plasma cell dyscrasias. J Clin Invest 1989; 84: 2008–2011.
Bataille R, Klein B . C-reactive protein levels as a direct indicator of interleukin-6 levels in humans in vivo. Arthritis Rheum 1992; 35: 982–984.
Moreau P, Harousseau JL, Wijdenes J, Morineau N, Milpied N, Bataille R . A combination of anti-interleukin 6 murine monoclonal antibody with dexamethasone and high-dose melphalan induces high complete response rates in advanced multiple myeloma. Br J Haematol 2000; 109: 661–664.
Rossi JF, Fegueux N, Lu ZY, Legouffe E, Exbrayat C, Bozonnat MC et al. Optimizing the use of anti-interleukin-6 monoclonal antibody with dexamethasone and 140 mg/m2 of melphalan in multiple myeloma: results of a pilot study including biological aspects. Bone Marrow Transplant 2005; 36: 771–779.
Dorjsuren D, Lin Y, Wei W, Yamashita T, Nomura T, Hayashi N et al. RMP, a novel RNA polymerase II subunit 5-interacting protein, counteracts transactivation by hepatitis B virus X protein. Mol Cell Biol 1998; 18: 7546–7555.
Parusel CT, Kritikou EA, Hengartner MO, Krek W, Gotta M . URI-1 is required for DNA stability in C. elegans. Development 2006; 133: 621–629.
Theurillat JP, Metzler SC, Henzi N, Djouder N, Helbling M, Zimmermann AK et al. URI is an oncogene amplified in ovarian cancer cells and is required for their survival. Cancer Cell 2011; 19: 317–332.
Yang H, Gu J, Zheng Q, Li M, Lian X, Miao J et al. RPB5-mediating protein is required for the proliferation of hepatocellular carcinoma cells. J Biol Chem 2011; 286: 11865–11874.
Adams J . The proteasome: a suitable antineoplastic target. Nat Rev Cancer 2004; 4: 349–360.
Mitsiades N, Mitsiades CS, Poulaki V, Chauhan D, Fanourakis G, Gu X et al. Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc Natl Acad Sci USA 2002; 99: 14374–14379.
Spooren A, Kooijman R, Lintermans B, Van Craenenbroeck K, Vermeulen L, Haegeman G et al. Cooperation of NFkappaB and CREB to induce synergistic IL-6 expression in astrocytes. Cell Signal 2010; 22: 871–881.
Yoon S, Woo SU, Kang JH, Kim K, Kwon MH, Park S et al. STAT3 transcriptional factor activated by reactive oxygen species induces IL6 in starvation-induced autophagy of cancer cells. Autophagy 2010; 6: 1125–1138.
Zhang Y, Bharadwaj U, Logsdon CD, Chen C, Yao Q, Li M . ZIP4 regulates pancreatic cancer cell growth by activating IL-6/STAT3 pathway through zinc finger transcription factor CREB. Clin Cancer Res 2010; 16: 1423–1430.
Ikebuchi K, Wong GG, Clark SC, Ihle JN, Hirai Y, Ogawa M . Interleukin 6 enhancement of interleukin 3-dependent proliferation of multipotential hemopoietic progenitors. Proc Natl Acad Sci USA 1987; 84: 9035–9039.
Catlett-Falcone R, Landowski TH, Oshiro MM, Turkson J, Levitzki A, Savino R et al. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity 1999; 10: 105–115.
Puthier D, Derenne S, Barille S, Moreau P, Harousseau JL, Bataille R et al. Mcl-1 and Bcl-xL are co-regulated by IL-6 in human myeloma cells. Br J Haematol 1999; 107: 392–395.
Hayashi T, Hideshima T, Anderson KC . Novel therapies for multiple myeloma. Br J Haematol 2003; 120: 10–17.
Hideshima T, Catley L, Raje N, Chauhan D, Podar K, Mitsiades C et al. Inhibition of Akt induces significant downregulation of survivin and cytotoxicity in human multiple myeloma cells. Br J Haematol 2007; 138: 783–791.
Brown CO, Salem K, Wagner BA, Bera S, Singh N, Tiwari A et al. Interleukin-6 counteracts therapy-induced cellular oxidative stress in multiple myeloma by up-regulating manganese superoxide dismutase. Biochem J 2012; 444: 515–527.
Demchenko YN, Kuehl WM . A critical role for the NFkB pathway in multiple myeloma. Oncotarget 2010; 1: 59–68.
Bharti AC, Shishodia S, Reuben JM, Weber D, Alexanian R, Raj-Vadhan S et al. Nuclear factor-kappaB and STAT3 are constitutively active in CD138+ cells derived from multiple myeloma patients, and suppression of these transcription factors leads to apoptosis. Blood 2004; 103: 3175–3184.
Hideshima T, Chauhan D, Richardson P, Mitsiades C, Mitsiades N, Hayashi T et al. NF-kappa B as a therapeutic target in multiple myeloma. J Biol Chem 2002; 277: 16639–16647.
Richardson PG, Sonneveld P, Schuster MW, Irwin D, Stadtmauer EA, Facon T et al. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N Engl J Med 2005; 352: 2487–2498.
Jakubikova J, Adamia S, Kost-Alimova M, Klippel S, Cervi D, Daley JF et al. Lenalidomide targets clonogenic side population in multiple myeloma: pathophysiologic and clinical implications. Blood 2011; 117: 4409–4419.
Patrawala L, Calhoun T, Schneider-Broussard R, Zhou J, Claypool K, Tang DG . Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2- cancer cells are similarly tumorigenic. Cancer Res 2005; 65: 6207–6219.
Hadnagy A, Gaboury L, Beaulieu R, Balicki D . SP analysis may be used to identify cancer stem cell populations. Exp Cell Res 2006; 312: 3701–3710.
Hirschmann-Jax C, Foster AE, Wulf GG, Nuchtern JG, Jax TW, Gobel U et al. A distinct "side population" of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci USA 2004; 101: 14228–14233.
Zhou S, Schuetz JD, Bunting KD, Colapietro AM, Sampath J, Morris JJ et al. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat Med 2001; 7: 1028–1034.
Dong LW, Hou YJ, Tan YX, Tang L, Pan YF, Wang M et al. Prognostic significance of Beclin 1 in intrahepatic cholangiocellular carcinoma. Autophagy 2011; 7: 1222–1229.
Wang Q, Tan YX, Ren YB, Dong LW, Xie ZF, Tang L et al. Zinc finger protein ZBTB20 expression is increased in hepatocellular carcinoma and associated with poor prognosis. BMC Cancer 2011; 11: 271.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81101790, 81370066, and 81372355), the Funds for Creative Research Groups of China (81221061), and the basic funding of Shanghai Changzheng Hospital of the Second Military Medical University (200901).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Ciliberto
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Fan, JL., Zhang, J., Dong, LW. et al. URI regulates tumorigenicity and chemotherapeutic resistance of multiple myeloma by modulating IL-6 transcription. Cell Death Dis 5, e1126 (2014). https://doi.org/10.1038/cddis.2014.93
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2014.93
Keywords
This article is cited by
-
ALCAM regulates multiple myeloma chemoresistant side population
Cell Death & Disease (2022)
-
R2TP/Prefoldin-like component RUVBL1/RUVBL2 directly interacts with ZNHIT2 to regulate assembly of U5 small nuclear ribonucleoprotein
Nature Communications (2017)
-
Melatonin improves age-induced fertility decline and attenuates ovarian mitochondrial oxidative stress in mice
Scientific Reports (2016)