Abstract
Cancer treatments induce cell stress to trigger apoptosis in tumor cells. Many cancers repress these apoptotic signals through alterations in the Bcl2 proteins that regulate this process. Therapeutics that target these specific survival biases are in development, and drugs that inhibit Bcl2 activities have shown clinical activity for some cancers. Mcl1 is a survival factor for which no effective antagonists have been developed, so it remains a principal mediator of therapy resistance, including to Bcl2 inhibitors. We used a synthetic-lethal screening strategy to identify genes that regulate Mcl1 survival activity using the pediatric tumor neuroblastoma (NB) as a model, as a large subset are functionally verified to be Mcl1 dependent and Bcl2 inhibitor resistant. A targeted siRNA screen identified genes whose knockdown restores sensitivity of Mcl1-dependent NBs to ABT-737, a small molecule inhibitor of Bcl2, BclXL and BclW. Three target genes that shifted the ABT-737 IC50 >1 log were identified and validated: PSMD14, UBL5 and PRPF8. The latter two are members of a recently characterized subcomplex of the spliceosome that along with SART1 is responsible for non-canonical 5′-splice sequence recognition in yeast. We showed that SART1 knockdown similarly sensitized Mcl1-dependent NB to ABT-737 and that triple knockdown of UBL5/PRPF8/SART1 phenocopied direct MCL1 knockdown, whereas having no effect on Bcl2-dependent NBs. Both genetic spliceosome knockdown or treatment with SF3b-interacting spliceosome inhibitors like spliceostatin A led to preferential pro-apoptotic Mcl1-S splicing and reduced translation and abundance of Mcl1 protein. In contrast, BN82865, which inhibits the second transesterification step in terminal spliceosome processing, did not have this effect. These findings demonstrate a prominent role for the spliceosome in mediating Mcl1 activity and suggest that drugs that target either the specific UBL5/PRPF8/SART1 subcomplex or SF3b functions may have a role as cancer therapeutics by attenuating the Mcl1 survival bias present in numerous cancers.
Similar content being viewed by others
Main
The Bcl2 family of proteins governs mitochondrial apoptosis in normal and cancer cells. Specific cellular stressors, including those initiated by anti-cancer therapy, activate select pro-death BH3-only proteins through diverse mechanisms.1 These proteins interact with Bcl2 family members at the outer mitochondrial membrane, where a subset, including Bid and Bim, can directly activate the obligate executioners Bak or Bax to commit the cell to death. Alternatively, activated BH3-only proteins may be sequestered within the hydrophobic pocket of the pro-survival proteins Mcl1, Bcl2, BclxL, Bclw or A1/Bfl, neutralizing their death signals.2 Neuroblastoma (NB) is a highly lethal childhood tumor that we have previously shown to be dependent on pro-survival Bcl2 family proteins to neutralize activated Bim and evade apoptosis.3, 4 They can be functionally classified into three categories: those dependent on Bcl2 for survival, those dependent on Mcl1, and post-therapy relapsed tumors with profound mitochondrial-apoptosis resistance. The small molecule ABT-737, an inhibitor of Bcl2, BclxL and Bclw, has low nanomolar potency against Bcl2-dependent cell lines and in vivo efficacy in pre-clinical models.3, 4 However, this agent does not antagonize Mcl1,5 which remains an important resistance mediator for Mcl1-dependent NBs and many other tumor histotypes. For this reason, there remains great interest in developing Mcl1 antagonists for clinical use.
Knocking down Mcl1 in Mcl1-dependent cancer cells restores sensitivity to ABT-737 confirming it as a principal survival factor,6 so we sought to use the specific activity of ABT-737 in a synthetic-lethal siRNA screen to identify targets that support Mcl1 activity. Mcl1 is unique among Bcl2 pro-survival family members in its short half-life (1.5–6 h) and myriad regulatory influences altering stability and function.7 Mcl1 transcription is induced downstream of cdks, Ras/Raf/Mek/Erk, PI3K/Akt and Jak/STAT3, and alternative splicing can generate multiple isoforms.8, 9 Mcl1 is further regulated post-translationally, with constitutive turnover occurring via ubiquitin-mediated degradation through the Huwe1 Hect E3-ligase.10 The E3-ligase, BTRC, operating downstream of AKT and GSK3β,11 and Fbw7, the substrate recognition subunit of the SCF complex, also function to degrade Mcl1.12 Conversely, deubiquitinases (DUBs) that selectively remove ubiquitin from substrates to promote their stability have been implicated in stabilizing oncoproteins, such as Mcl1.13 Usp9x has been proposed as a specific Mcl1 DUB in lymphomas and multiple myeloma, and interfering with its activity restores sensitivity to ABT-737,14 though no such correlation has been identified in NB.
Given the extensive regulation of Mcl1 by the ubiquitin–proteosome system, we hypothesized that a specific DUB sustained Mcl1 activity in NB. We therefore chose an siRNA library targeting DUBs for our screen and identified gene targets that sensitize cells to ABT-737. Unexpectedly, we validated that the proteosomal lid component, PSMD14, and multiple components of the splicosome (PRPF8, UBL5, and SART1) promote Mcl1 pro-survival activity. The latter were included in the library as they contain DUB or ubiquitin-like motifs, although their Mcl1 effects are independent of canonical DUB activities. Inactivating these components by RNAi or biochemical inhibition causes pro-apoptotic splicing changes in Mcl1, reduced Mcl1 translation and restored sensitivity of Mcl1-dependent NBs to ABT-737. Collectively, these data identify targets for restoration of apoptotic signaling in Mcl1-dependent cancers by exploiting its highly regulated nature, specifically in antagonizing spliceosome-mediated events.
Results
An siRNA screen identifies PRPF8, UBL5 and PSMD14 as targets that are synthetic-lethal with Bcl2 antagonists in Mcl1-dependent cancers
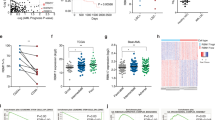
We used an siRNA library to target 98 DUBs in two cell lines dependent on Mcl1 for survival: IMR5 and NLF.3 Though both express Bcl2 protein, they neutralize Bim exclusively through Bim:Mcl1 binding and are resistant to the Bcl2 antagonist ABT-737 in vitro and in vivo.3, 4 Direct knockdown of Mcl1 sensitizes these cells to ABT-737 (IC50 2 and 30 nM, respectively), >100-fold less than their IC50 to ABT-737 without Mcl1 knockdown. We screened in the presence of 200 nM ABT-737 as this concentration was >10-fold less than the IC50 in parental cells and ∼10-fold greater than the IC50 following Mcl1 knockdown, demonstrating maximally divergent cytotoxicity (Figure 1a). Using a Z-score of <−1.5 in cells co-treated with ABT-737 and siRNA and a ratio of (ATP-ABT737/ATP-vehicle) <0.80 to define our hit threshold, PSMD14, PRPF8, UBL5 and UCHL3 were identified as Mcl1 activators in IMR5 and PSMD14, PRPF8, and USP17 in NLF (Figures 1b and c and Supplementary Table S1). USP9X had Z-scores of 0.40 in IMR5 and −0.10 in NLF, and Usp9x protein levels did not correlate with Mcl1 protein or activity. Indeed, Usp9x activity had not been identified in neural tissues or NB cell lines, in contrast with hematolymphoid tissues.15
Primary siRNA screen. (a) NB cells known to be Bcl2 dependent (SMS-SAN) are sensitive to ABT-737, while Mcl1-dependent NB cells (IMR5 and NLF) are ABT-737 resistant due to Mcl1 activity, unless Mcl1 is knocked down using siRNA. The ABT-737 concentration used in our primary screen (200 nM; shown) provides maximally divergent responses in the presence or absence of Mcl1 activity. Primary screen results are shown for (b) IMR5 and (c) NLF cells using pooled siRNAs targeting each of 98 DUBs and treated with 200 nM ABT-737 or vehicle control (screen hits are labeled and have Z-score <−1.5 and viability ratio with ABT-737 to without ABT-737 of <0.80); grey bars show ATP content following siRNA alone, red bars show percentage of change in ATP with siRNA in the presence of ABT-737. (d–f) ABT-737 cytotoxicity was measured following siRNA knockdown of screen hits for verification. Consistent with our primary screen results, siRNA targeting PSMD14 and PRPF8 sensitized both IMR5 and NLF cells to ABT-737, while siUBL5 sensitized only IMR5 cells. Knockdown of neither target gene altered ABT-737 sensitivity of SMS-SAN cells that are not Mcl1 dependent. ATP content is used as a cell viability surrogate; siSham, non-targeting (control) siRNA; error bars, S.E.M. Data represent two independent experiments except for b and c, which were single primary screens
We verified screen results with siRNA knockdown of each target gene markedly reducing the IC50 to ABT-737 except for USP17, which was not studied further (Figures 1d–f and Table 1). We next measured the ABT-737 IC50 with each individual siRNA from the pool targeting each gene, and multiple siRNA constructs targeting PRPF8, PSMD14 and UBL5 each demonstrated a >10-fold reduction in ABT-737 IC50 supporting on-gene effects (Table 1). Only one UCHL3-targeted siRNA construct shifted the IC50 so UCHL3 was not studied further. We measured the ABT-737 IC50 across NB cell lines verified as Bcl2 dependent (SMS-SAN) or therapy resistant (SK-N-AS and BE2C, established at the time of relapse).3 The notable reduction in ABT-737 IC50 was confined to tumor cells functionally dependent on Mcl1 (with the exception that PSMD14 knockdown decreased the IC50 to ABT-737 in therapy-resistant BE2C cells;3, 4 Table 1). Thus, UBL5, PRPF8 and PSMD14 were validated hits, with PSMD14 and PRPF8 confirmed in both cell lines.
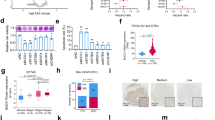
To further validate on-target activities, we performed rescue experiments expressing each gene with a translationally silent mutation rendering it siRNA resistant. Expressing knockdown-resistant pUBL5 (P<0.01) or pPSMD14 (P<0.01) rescued cells from siRNA-mediated ABT-737 sensitization, compared with LacZ control transfection (Figure 2). We were unable to stably express pPRPF8, so rescue of its knockdown phenotype could not be tested. As we initially sought a DUB activity that sustained Mcl1, we assessed whether these genes functioned through ubiquitin-mediated mechanisms. We engineered an siRNA-resistant PSMD14 mutating two critical JAMM motif residues, H113A and H115A (pPSMD14-H>A), that failed to rescue the phenotype of PSMD14 knockdown, supporting that its protease activity is required to promote Mcl1 function (Figure 2c). In contrast, UBL5 is a ubiquitin-like modifier that lacks the C-terminus diglycine (GG) by which ubiquitin is bound to its substrates. Instead, it has a dityrosine before a terminal non-conserved residue.16 Similar ubiquitin-like modifications have been implicated in regulating Bcl2 family proteins,17 but an siRNA-resistant UBL5 that lacked the terminal YYQ residues (pUBL5-Cterm) did rescue cells from siRNA knockdown (P=0.03) similar to wild-type UBL5, indicating that C-terminal conjugation to substrates is unlikely to be its primary mechanism for Mcl1 antagonism (Figure 2).
Rescue with siRNA-resistant wt and mutant UBL5 and PSMD14. IMR5 and NLF cells were transfected with plasmids expressing siRNA-resistant wt UBL5 (pUBL5) or conjugation-deficient UBL5 (pUBL5-Cterm; lacking terminal YYQ residues) or siRNA-resistant wt PSMD14 (pPSMD14) or DUB-protease inactivated H113A/H115A mutant (pPSMD14-H->A), respectively. Cells were assessed for ABT-737 sensitivity following siRNA knockdown and compared with lacZ control transfectants (pLacZ). (a) Expression of siRNA-resistant wt or conjugation-defective UBL5 rescued cells from ABT-737 cytotoxicity supporting on-gene effects for Mcl1 antagonism that do not require a ubiquitylation-like activity for UBL5. (c) Conversely, expression of siRNA-resistant wt PSMD14 rescued cells from ABT-737 cytotoxicity but expression of a DUB-protease-deficient PSMD14 did not, suggesting Mcl1 antagonism activity is mediated by PSMD14 and requires its protease domain. (b, d) Expression of each gene was measured by qPCR and normalized to HPRT and to the expression in LacZ control cells treated with non-targeting siRNA for comparison of target-gene modulation with functional effect. ATP content is used as a cell viability surrogate; wt, wild-type; siSham, non-targeting (control) siRNA; error bars, S.E.M. Data are representative of at least two independent experiments
Spliceosome complex components are potential regulators of Mcl1
The Saccharomyces cerevisiae homolog of UBL5, Hub1, functions within a spliceosome complex with Snu66 and Prp8 (the yeast homolog of PRPF8), binding via a novel HIND interaction domain independent of the terminal dityrosine.15 This complex does not effect global spliceosome activities in yeast but is required for alternative (in distinction to canonical) 5′-splice site usage.18 As two of these complex members were hits in our screen (UBL5/Hub1 and PRPF8/Prp8), we knocked down SART1 (the human homolog of snu66) and this similarly sensitized IMR5 cells to ABT-737 (IC50 916 nM versus 6.8 μM for sham siRNA, Supplementary Figure S1A). Simultaneous knockdown of UBL5, PRPF8 and SART1 nearly phenocopied direct Mcl1 knockdown, whereas this triple knockdown had no effect in SMS-SAN cells that are Bcl2 dependent (Supplementary Figure S1B). These data support a novel role for this spliceosome complex in mediating Mcl1 activity.
Mcl1 protein expression was reduced by knockdown of UBL5, SART1 or PRPF8 in Mcl1-dependent cells (in contrast to PSMD14 knockdown that led to Mcl1 accumulation, Figure 3a). Given a postulated role for the spliceosome in this process, we assessed Mcl1 mRNA splicing. MCL1-L is the ubiquitous pro-survival isoform, MCL1-S is generated by exon skipping and contains a BH3 death domain and MCL1-ES has a novel intron in the first exon of MCL1-L but is not expressed in NB.8, 9 Despite MCL1-L protein reduction with spliceosome component knockdown, MCL1-L mRNA was not consistently reduced, suggesting transcriptional repression is not a major effector (Figure 3b). Knockdown of spliceosome complex members increased MCL1-S abundance and the MCL1-S/MCL1-L ratio as confirmed by both RT-PCR and QPCR (Figures 3c and d), and as Mcl1-S is reported to be pro-apoptotic, we tested whether this level of MCL1-S expression phenocopies spliceosome knockdown. First, we expressed Mcl1-S using a tetracycline-inducible construct, and there was a trend to an inverse correlation between ABT-737 sensitivity (IC50) and MCL1-S abundance across subclones (r=−0.78, P<0.07; Figure 4), yet even those expressing MCL1-S comparable to spliceosome-component knockdown (see Figures 3c and 4d) did not fully restore ABT-737 sensitivity (IC50 reduced only to 1.4 μM). We also used morpholino oligonucleotides to induce exon skipping and promote Mcl1-S splicing.19 Again a dose–response effect was seen (more notable in IMR5) that again led to only modest reductions in ABT-737 IC50 (>1 μM; Figure 4). Therefore, genetic knockdown of UBL5/PRPF8/SART1 increased Mcl1-S, but our data suggest that this is not the principal effector for Mcl1 antagonism but that reduced Mcl1-L protein is the major contributor.
Effects of knockdown of identified hits on Mcl1 mRNA and protein. (a and b) IMR5 and NLF cells were transfected with siRNAs to the indicated genes for 72 h, and protein and mRNA were harvested. PSMD14 knockdown leads to increased Mcl1 protein (relative to β-tubulin loading control) and mRNA, while knockdown of the spliceosome components PRPF8, UBL5 or SART1 leads to (a) reduced Mcl1 protein but with (b) increased MCL1-L mRNA for most, rather than reduced mRNA. (c and d) Complementary QPCR and RT-PCT methods concordantly demonstrate increased splicing toward MCL1-S following knockdown of spliceosome complex members PRPF8, UBL5 or SART1 leading to an increased MCL1-S/MCL1-L ratio. siSham, non-targeting (control) siRNA; error bars, S.E.M. ND, not determined as siMcl1 led to loss of all viable cells. *, P-values ≤0.05 versus siSham. Data are representative of at least two independent experiments
Impact of MCL1-S expression on ABT-737 sensitivity. MCL1-S was expressed from the pDest30 vector in IMR5 cells and subclones identified by serial dilution. (a) ABT-737 cytotoxicity for six independent subclones was measured and (b) correlated with their MCL1-S expression by RT-PCR. Similarly, ABT-737 cytotoxicity curves were obtained using (c) IMR5 and (e) NLF cells by treating with increasing concentration of morpholino oligonucleotides (designated Mcl1) to induce exon skipping toward MCL1-S expression and (d and f, below) correlated with their MCL1-S expression. Although there was a linear correlation between increasing MCL1-S expression and decreasing ABT-737 IC50 (for IMR5 pMcl1-S, panel a: r=−0.78 and P<0.07; for IMR5-morpholino, panel c: r=−0.95 and P<0.01; for NLF-morpholino, panel e: r=−0.76 and P<0.08), the magnitude of this effect was marginal (lowest IC50 of 1.3 μM) by comparison with the reduced ABT-737 IC50 seen with spliceosome component knockdown (IC50<100 nM) despite similar induced MCL1-S splicing. ATP content is used as a cell viability surrogate; siSham, non-targeting (control) siRNA; error bars, S.E.M. Data are representative of at least two independent experiments
In addition to Mcl1 itself, regulators of Mcl1 might be alternatively spliced to alter Mcl1 activity, so we assessed splicing in a panel of such genes in response to spliceosome knockdown: BCL2L1 (Bclx), BCL2L11 (Bim), BAX, MAPK8 (Jnk), EPHB2 (Erk), GSK3B, HUWE1, BTRC, USP9X, EIF4E, AKT1, and STAT3. Similar to Mcl1, the predominant change was an increase in exon skipping (for BAX, GSK3B, HUWE1 and BTRC), while there was increased alternate splice donor site usage for BCL2L1 to generate Bclx-S preferentially over Bclx-L (Figure 5a). The remaining genes had no splice alterations identified. Notably, no non-canonical 5′-splice donor site splice alterations, as regulated by Hub1/Prp8/Snu66 activities in yeast, were seen. Overall, splicing changes were most pronounced with BTRC, reducing the full-length isoform while increasing exon 2 skipped product, and generating novel exon 2–3 and exon 2–4 skipped products. None of these alternatively spliced products (that generate in frame deletions that skip amino acids 17–52, 17–78 and 17–108, respectively) have been reported to antagonize Mcl1,20 so their effect on Mcl1 regulation is unclear.
Splicing of candidate Mcl1 regulators and impact of knockdown on Mcl1 protein stability. (a) RT-PCR with primers recognizing major splice isoforms for the indicated genes (see Materials and Methods) demonstrates an increase in alternate donor site usage in BCLX, generating the pro-apoptotic BCLX-S, and in exon skipping in BAX, GSK3B, HUWE1 and BTRC with knockdown of spliceosome components SART1, UBL5 (minor) and/or PRPF8; (b) Mcl1 protein stability was assessed following siRNA knockdown of UBL5, PRPF8 and PSMD14. Cells were harvested after treatment for the indicated period with cyclohexamide to inhibit protein synthesis and blotted for Mcl1 and tubulin. siSham=non-targeting (control) siRNA; FB, fetal brain
We were unable to forcibly overexpress these splice-altered BTRC isoforms at the protein level to directly assess their impact on Mcl1 stability or activity. To indirectly assess whether BTRC or an unidentified regulator decreased Mcl1 stability through altered post-translational modification, we measured the survival of Mcl1 protein following inhibition of protein synthesis 48 h after siRNA knockdown. Knockdown of PSMD14 consistently increased Mcl1 half-life consistent with its function in the proteasome; however, knockdown of neither UBL5 nor PRPF8 shortened Mcl1 half-life (Mcl1-L or Mcl1-S), suggesting that decreased protein stability is not the major determinant of loss of Mcl1 protein (Figure 5b). Collectively, our data support that spliceosome knockdown leads to increased splicing to Mcl1-S and associated reduced Mcl1-L translation, without reducing transcription or protein stability. As splicing is required for the efficient transport of mRNA to the cytosol for translation, it is possible that altered spliceosome activities lead to reduced Mcl1-L protein through inhibited mRNA trafficking and translation.21
Small molecule spliceosome complex inhibitors phenocopy Mcl1 antagonism
Ubl5, Prpf8 and Sart1 copurify with the U5-U4/U6 tri-snRNP complex of the spliceosome that is recruited to join U1 and U2 snRNP complexes that recognize splice donor and acceptor sites.22, 23, 24 Whether Ubl5/Prpf8/Sart1 have restricted roles or provide target selectivity within the spliceosome is unknown, and no inhibitors of this subcomplex have been identified. However, inhibitors of SF3b, a component that directly interacts with Prpf825 and inhibits the immediate upsteam U2 snRNP complex, have been developed.26 Knockdown of SF3B1, the SF3B member most highly expressed in NBs, sensitized Mcl1-dependent IMR5 and NLF cells to ABT-737 (IC50 of 64 and 276 nM, respectively) and markedly skewed Mcl1 splicing toward the pro-apoptotic Mcl1-S isoform, analogous to Ubl5, Prpf8 and Sart1 knockdown (Figures 6a–c). Biochemical inhibition of SF3b by spliceostatin A (SSA), a methylated derivative of FR901464,26 resulted in near-abolished Mcl1-L expression and, importantly, phenocopied direct Mcl1 knockdown with respect to ABT-737 sensitization (IC50 to ABT-737 of 6.8 and 28.5 nM in IMR5 and NLF, respectively; Figures 6d and e). SSA-mediated resensitization to ABT-737 was seen not only in the Mcl1-dependent subset but also in therapy-resistant cells (with repressed mitochondrial signaling) that utilize Mcl1 to sequester activated Bim,4 as SSA increased sensitivity to ABT-737 in BE2C cells (IC50 13 nM) and, to a lesser degree, in SK-NAS cells (Figures 6i and j). This suggests that spliceosome antagonists may have utility in the relapsed setting for chemoresistant tumors that depend on Mcl1. To see whether there was specificity within the spliceosome for this activity, we investigated BN82865, a napthoquinone that blocks the second transesterification step acted upon complex C*, a late step in spliceosome processing.27 Unlike SSA and other U2 antagonists like Meayamycin B, BN82865 did not alter Mcl1 activity or ABT-737 sensitivity in Mcl1-dependent cells (Supplementary Figure S2), despite inhibiting the spliceosome complex that still included Ubl5, Prp8 and Sart1 supporting that earlier spliceosome inhibition correlated with Mcl1 antagonism.27
Effect of genetic and biochemical inhibition of the spliceosome component SF3B1. (a) IMR5 and (b) NLF cells were transfected with siRNA targeting SF3B1, MCL1 or sham control. (c) SF3B1 knockdown markedly increased ABT-737 cytotoxicity (P<0.01) while increasing MCL1-L (modestly) and MCL1-S (markedly) as measured by QPCR. Biochemical inhibition of SF3B1 was tested using SSA (10 nM) or vehicle control (MeOH) and assessing ABT-737 cytotoxicity in diverse NB cell lines, including (d) IMR5 and (e) NLF —both Mcl1 dependent; (h) SMS-SAN —Bcl2 dependent; and (i) SK-N-AS and (j) BE2C—both therapy resistant. Cells were treated with either siRNA reagents or SSA or vehicle for 24 h before ABT-737 exposure for 48 h. The impact of SSA on Mcl1 splicing is shown for IMR5 cells treated with 10 nM SSA, with Mcl1-S and Mcl1-L isoforms assessed at the mRNA level (detected at 6 h using RT-PCR; panel f, top) and protein level (detected at 8 h by IB; panel f, bottom) and (g) confirmed using complementary QPCR. SSA markedly skews Mcl1 splicing toward Mcl1-S and reduces Mcl1-L protein. ATP content is used as a cell viability surrogate; siSham, non-targeting (control) siRNA; error bars, S.E.M.; *P<0.05. Data are representative of at least two independent experiments (run in technical triplicates for panels c and g)
SSA induced a near-complete shift from Mcl1-L to Mcl1-S isoforms, yet although Mcl1-S mRNA exceeded those seen with spliceosome knockdown, this was not reflected in significantly increased Mcl1-S protein (Figures 3 and 6f). Instead, as with spliceosome or direct Mcl1 knockdown, ABT-737 sensitization directly paralleled the reduction in Mcl1-L protein. We found that SSA alters the splicing of the Mcl1 regulators BTRC and GSK3B in a similar pattern as SART1 and PRPF8 knockdown, but these effects are less pronounced compared with Mcl1 splicing changes, and there are no effects on the splicing of BCL2L1 (Bclx), BAX or HUWE1 (Supplementary Figure S3). The effect of SSA therapy on Mcl1 splicing was not restricted to NB as treatment of HeLa, PANC-1 and MCF7 cells (that express Mcl1 and are ABT-737 resistant) altered the splicing of Mcl1 to a nearly equivalent degree (Supplementary Figure S4). The lack of sensitization to ABT-737 in these cells supports that functional studies rather than Mcl1 protein expression alone are required to define cells with Mcl1 dependence.
Expression of splicing components correlates with poor outcome NB
Mcl1 activity provides a survival bias in cancer and has been associated with therapy resistance in NB.6 We therefore correlated the expression of spliceosome subcomplex genes with prognostic variables and clinical outcome in NBs (r2.amc.nl;28). Using an expression data set of 649 NBs with associated prognostic information, the poor prognosis variables of age >18 months at diagnosis, stage 4 disease and MYCN amplification were all highly associated with high expression of SART1, UBL5, PRPF8 and SF3B1 at P<0.001, except for PRPF8 and MYCN amplification (P=0.11; Supplementary Figure S5). An independent set of 88 NBs in which survival outcome was also known showed an association between high SART1 and PRPF8 with inferior event-free survival (EFS) and overall survival and with SF3B1 with EFS alone (Supplementary Figure S5). These correlations are consistent with a role for this spliceosome subcomplex in promoting Mcl1 activity, and survival bias, in NB. There was no correlation between Mcl1 mRNA expression and expression of any of these spliceosome components, consistent with our data that these are working through a post-translational mechanism and not directly on Mcl1 transcription.
Discussion
Apoptosis suppression is one of the hallmarks of cancer.29 Alterations in Bcl2 family proteins commonly mediate this, so numerous agents that target Bcl2 family activities are in clinical development.30 This includes the ABT-737 homolog, Navitoclax (Abbvie), a Bcl2, BclxL and BclW inhibitor with demonstrated activity in chronic lymphocytic leukemia31 that is being studied in numerous clinical trials (clinicaltrials.gov). Navitoclax does not antagonize Mcl1, however, and this protein remains a major contributor to de novo and acquired resistance.32 Therefore, there is great interest in identifying antagonists of Mcl1’s pro-survival activity to potentially revert therapy resistance in Mcl1-dependent cancers or to synergize with Bcl2 antagonists by preventing emergence of Mcl1-mediated resistance.
Cancer cell dependence on specific Bcl2 homologs is defined functionally, as redundancy and extensive post-translational regulation make predictions from their expression levels alone difficult. For many tumors, their selective survival dependence reflects tonic Bim sequestration by specific Bcl2 pro-survival proteins.4, 33 Many NBs are Mcl1 dependent and resistant to Bcl2 antagonists, demonstrating Bim sequestration by Mcl1.4, 6 Knockdown of Mcl1 restores sensitivity to chemotherapy and to ABT-737 in this subset both in vitro and in vivo.4 We exploited this dominant survival bias, and the highly regulated nature of Mcl1, using a focused siRNA synthetic-lethal screen that was highly specific, with all targets with a Z-score <−2 validated as having effects on Mcl1 expression and activity. Our screen focused on DUBs to identify a DUB that rescues Mcl1 from proteosomal degradation in NB, analagous to USP9X in lymphoma.14 We identified no such enzyme; however, it is possible that not all genes in our screen achieved adequate knockdown, particularly for proteins with a longer half-life, or that redundant DUB proteins promote Mcl1 activity in neural cells. We identified PSMD14, a component of the 19S proteasomal lid that has DUB activity, as a validated target whose knockdown leads to stabilization of Mcl1-L protein and paradoxical sensitization to ABT-737. Two additional 19S proteasome lid proteins have attributed DUB activity, Uchl5 and Usp14,34 yet neither were hits in our screen, suggesting that there may be selectivity in substrate selection within this complex (Supplementary Table S1). Further work in this area is warranted as more selective proteasome inhibitors are now under development, including those that target 19S DUB activities,34 and a less toxic Mcl1-specific inhibitor may be achievable.
Two validated targets, PRPF8 and UBL5, are components of the spliceosome and we focused on them as potential novel Mcl1 regulators, especially as Mcl1 has both pro- and anti-apoptotic isoforms making this is an attractive mode of control.8, 9 In yeast, loss of the UBL5 homolog Hub1 results in defective splicing at non-canonical 5′ splice donor sites. The regulation of alternative splicing is substantially more complex in humans; however, Ubl5 and Sart1 physically interact25 and co-localize in nuclear speckles18 in human cells and associate with Cajal bodies, the nuclear subdomains where splicesome components are assembled.35 This suggests that the subcomplex may serve similar activities. Our data that knockdown of UBL5 or SART1, the homolog of its yeast binding partner snu66, both induce alternative splicing confined to exon skipping and alternative donor site usage supports this conservation. Still, transcriptome-wide approaches will be important to broadly characterize the specific splicing alterations mediated by these proteins across diverse normal and malignant tissues.
Interestingly, Mcl1 survival activity antagonism was limited to perturbations predicted to impair spliceosome assembly and was not seen with an inhibitor of the downstream second transesterification reaction. Sf3b proteins are components of the U2 complex that initially assembles on pre-mRNA to form complex A. Prpf8 is a core component of the U5 complex that associates with the U4/U6 complex (including Sart1 and thus likely Ubl5) and joins complex A on pre-mRNA to form complex B during splicing (Figure 7).36 Roybal and Jurica37 reported that SSA slows the transition from complex A to complex B with accumulation of an A-like complex with SSA treatment of nuclear extracts. Further, this complex formation was dependent on the 5′-splice donor site and branch point, but not the 3′ acceptor site. We hypothesize that knockdown of components of complex B (PRPF8, UBL5 and SART1) may similarly slow this transition and explain the similar phenotype. Members of both complexes A and B were previously identified as Mcl1 splice regulators using an Mcl1-splice reporter construct in HeLa cells, in a targeted screen of genes that regulate BclXL splicing.38 PRPF8 was a hit, but neither UBL5 nor SART1 were tested. The SR proteins SRSF1 and SRSF3 that regulate early spliceosome formation and splice site recognition were implicated in Mcl1 splicing in that study (as has been SRSF539). As with our findings, genetic knockdown of these genes led to a marked decrease in Mcl1-L protein but no change in protein stability or Mcl1-L mRNA (and variable increases in Mcl1-S mRNA) supporting an impact on Mcl1 translation.39 The mechanisms for this in NB require further studies but may include reduced recruitment of mTORC1 to the pre-mRNA, which has been linked to decreased translation.40, 41
Spliceosome activities and Mcl1 regulation summary. The U1 spliceosome subunit is recruited to the 5′ splice donor site at introns to form complex E. The U2 subunit (including SF3B) is subsequently recruited to form complex A, followed by recruitment of the U5-U4/U6 subunits (that includes Prpf8, Ubl5 and Sart1) generating complex B. The first transesterification reaction then occurs generating a lariat and releasing the U1 and U4 subunits. The second transesterification reaction occurs generating the mature mRNA by ligating the adjacent exons and releasing the remaining spliceosome subunits for recycling and the excised intron for degradation. SSA binds SF3B and delays progression from complex A to complex B, and SSA treatment or knockdown of the complex B subunits PRPF8, UBL5 or SART1 increase pro-apoptotic Mcl1-S splicing, decrease translation of antiapoptotic Mcl1-L protein and blocks the Mcl1 survival bias in Mcl1-dependent tumors, restoring sensitization to Bcl2 antagonists, such as ABT-737. BN82685 inhibits the second transesterification reaction and does not share this phenotype
Our work identifies spliceosome inhibition as a potential therapeutic target to antagonize Mcl1 in NB. Alternative splicing may similarly regulate Bcl2 family proteins in diverse tissues,42, 43 as supported by our finding that SSA induces Mcl1-S splicing in carcinoma cell lines of diverse origin. However, alternative splicing shows tissue specificity and is particularly important in neural development, with many neuronal-restricted splice isoforms identified,44, 45 so the mechanisms here could be restricted to neural tumors. Indeed, zebrafish expressing a hypomorphic SF3B146 gene or truncated PRPF847 demonstrate selective defects in neural crest development and neuronal apoptosis, respectively, while constitutional PRPF8-inactivating mutations in humans are causative of retinitis pigmentosa, a disease of the neural crest-derived retina.48 However, a genome-scale siRNA knockdown screen using multiple myeloma, a hematolymphoid tumor with an Mcl1 survival dependence,49 also identified three splicing components (PRPF8, SF3A1 and SNRPA1) as demonstrating selective lethality, and spliceosome components were the only hits with higher activity than Mcl1 knockdown itself.50 In this study, seven proteasome targets were also identified (though not PSMD14) supporting that, while the specific regulatory proteins identified may differ across cancer types, the core regulatory pathways may be conserved in Mcl1-dependent cancers.
There is much interest in spliceosome inhibitors as anti-cancer therapeutics,51 both because spliceosome components (including PRPF8 and SF3B1) have been identified as driver genes in hematological cancers52 and because cancer cells may have a unique dependence on spliceosome activities.53, 54 Spliceosome inhibitors, including natural products such as FR901464 and pladienolides, and synthetic analogs, such as SSA and meayamycin B, have potent anticancer activities but the basis for their therapeutic index remains unclear.55, 56 Recently, meayamycin B was shown to alter Mcl1 splicing and restore sensitivity to ABT-737 in non-small cell lung cancer cells, although this occurred via reduced Mcl1-L mRNA.57 We propose that one mechanism of selectivity is by decreasing the expression and function of Mcl1-L, allowing the activated BH3-only pro-death proteins expressed in tumor cells to induce apoptosis while sparing normal tissues, which are not primed for death by tonically activated BH3-only proteins. In future work, it will be important to identify whether the effect of spliceosome inhibitors on Mcl1 can be achieved with selective inhibition of splicing activities, as suggested by our finding that the Ubl5 and Sart1 are integral to this activity or requires more global splicing disruption.
Materials and Methods
Cell lines
NB cell lines previously shown to be Mcl1 dependent (IMR5,58 NLF), Bcl2 dependent (SMS-SAN, NB-164359) and resistant (SK-N-AS, BE2C) were used. Cells were grown in RPMI-1640 (Life Technologies, Grand Island, NY, USA) supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 U/ml of penicillin and 100 mcg/ml streptomycin. Tissue culture was at 37 °C in a humidified atmosphere of 5% CO2. All cell lines were confirmed as unique using STR-based genotyping (AmpFISTR, Life Technologies) and identity-confirmed using the COG cell line genotype database (www.cogcell.org). All cell lines routinely tested negative for mycoplasma contamination.
Optimization of transfection conditions
Transfection conditions were optimized for each cell line to achieve >70% knockdown for reporter genes (PPIB and GAPDH) with >80% viability using the Dharmacon (Pittsburgh, PA, USA) RTF Optimization Protocol 1. Dharmacon Cell Culture Reagent was used to dilute DharmaFect 1–4 transfection reagents with three-fold serial dilutions to concentrations ranging from 0.025 to 2.025 μl of Dharmafect per 25 μl of final suspension. White opaque 96-well plates (Fisher no. 08–771–26, Pittsburgh, PA, USA) prespotted with 1.25 pM of siRNA directed against cyclophilin B or a non-targeting control pool (Dharmacon RTF plates) were rehydrated with 25 μl of the dilutions of each transfection reagent in DCCR and allowed to sit at room temperature for 30 min. Cells were taken up with trypsin and counted using a hemocytometer. A suspension of 10 000 and 20 000 cells/100 μl was made using antibiotic-free complete media. A hundred microliters of this suspension was dispensed into each well on the siRNA plate. Cells were cultured for 48 h and then viability read using Cell Titer Glo (Promega, Madison, WI, USA) according to the manufacturers’ instructions in duplicate. RNA was extracted from the wells containing siRNA targeting Cyclophilin B and non-targeting controls and reverse transcription performed per the manufacturers’ protocol using the Ambion Cells-to-CT kit (Life Technologies), including an in-solution DNAse I digestion step. Knockdown was measured using Q-RT-PCR. Transfection conditions were chosen that resulted in >80% viability relative to untreated control and >50% knockdown of the target gene. Cell lines that had no such conditions identified (NB-1643) were deemed untransfectable in this system and omitted from further screening.
Primary siRNA screening
Duplicate pre-spotted 96-well plates containing 6.25 pM of siRNA targeting the deubiquiting enzymes (Dharmacon RTF ON-Target Plus library) were rehydrated with 25 μl of DharmaFect Cell Culture Reagent containing 0.075 μl DharmaFect 1 per well. After a 30 min incubation, 10 000 cells in 100 μl antibiotic-free otherwise complete RPMI was added to each well. After 24 h of incubation in standard conditions, 5 μl of antibiotic-free RPMI with or without ABT-737 at a concentration to give a final ABT-737 concentration of 200 nM was added to each well. Plates were incubated for an additional 48 h, and then ATP content was measured using Cell Titer Glo (Promega) per the manufacturers’ instructions. ATP content in each well was calculated by normalizing the luminesce reading to a standard dilution of ATP (arbitrary units). Z-scores were calculated for each well by subtracting the measured ATP content from the mean ATP content for all experimental wells for the cell line and dividing by the S.D. of the ATP content for all experimental wells. Hits were defined as having a Z-score <−1.5 and a ratio of (ATP: treated with ABT-737)/(ATP: treated with vehicle) <0.80.
Cytotoxicity assays
For siRNA experiments, OnTarget Plus siRNA (Dharmacon) was rehydrated with siRNA Resuspension Buffer (Dharmacon). The siRNA sequences used for rescue experiments were selected from the pool of four as the siRNA having the greatest sensitization to ABT-737. The following siRNA were used: PSMD14 5′-GGCAUUAAUUCAUGGACUA-3′, PRPF8 5′-GCAGAUGGAUUGCAGUAUA-3′ and UBL5 5′-GACCUUAAGAAGCUGAUUG-3′. For all other experiments, the pool of four siRNA targeting each gene was used and compared with ON-TARGETplus Non-targeting Control Pool (Dharmacon), a pool of four scrambled siRNA that do not target any human mRNA (siSham). In all, 6.25 pM of siRNA and 0.075 μl DharmaFect 1 were combined in 25 μl of DharmaFect Cell Culture Reagent per well. After 30-min incubation, 10 000 cells in 100 μl of antibiotic-free complete media was added to each well. After a 24-h incubation, ABT-737 was added in 5 μl of antibiotic-free complete medium. Forty-eight hours later, ATP content was measured using Cell Titer Glo (Promega) per the manufacturer’s instructions. For experiments using chemical spliceosome inhibitors, cells were plated at the above densities and allowed to settle overnight. Cells were treated with SSA for 24 h (a kind gift of Minoru Yoshida, Riken Institute, Wako, Japan), Sudemycin C1 or E for 48 h (kind gifts of Thomas Webb, St. Jude Children’s Research Center, Memphis, TN, USA), Meayamycin for 48 h (a kind gift of Kazunori Koide, University of Pittsburgh, Pittsburgh, PA, USA) or vehicle control and ABT-737 at the reported concentration and then ATP content was measured as above.
QPCR
Cells transfected as for cytotoxicity assays were cultured for 48 h. RNA was extracted from the wells containing siRNA targeting Cyclophilin B and non-targeting controls and reverse transcription performed per the manufacturers’ protocol using the Ambion Cells-to-CT kit for siRNA knockdown in 96-well plates, including an in-solution DNAse I digestion step. For splicing interrogation, RNA from cells transfected in six-well plates was collected per the manufacturer’s protocol using Qiagen’s RNeasy kit (Qiagen, Venlo, Netherlands), including an on-column DNAse digestion step. cDNA was made using the SuperScript III First-Strand Synthesis SuperMix per the manufacturor’s protocol with 1 μg of RNA and then diluted 1 : 10 before PCR. Q-RT-PCR was performed on an Applied Biosystems 7900-HT instrument, using primer probes for Mcl1, UBL5, PSMD14, PRPF8, SART1, GapDH, HPRT, and the TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA, USA). Relative mRNA was quantified using standard curves constructed from fetal brain RNA and normalized to total cellular mRNA content using HPRT as a neural housekeeping gene.
RT-PCR
For siRNA experiments, cells transfected as for cytotoxicity assays were cultured for 48 h. For chemical spliceosome inhibitor experiments, cells were plated, allowed to settle overnight and then treated with drug or vehicle control for 6 h. RNA from cells transfected in six-well plates was collected per the manufacturer’s protocol using Qiagen’s RNeasy kit, including an on-column DNAse digestion step. cDNA was made using the SuperScript III First-Strand Synthesis SuperMix (Invitrogen, Grand Island, NY, USA) per the manufacturer’s protocol with 1 μg of RNA and then diluted 1 : 10 before PCR. PCR using primers spanning splice junctions (Supplementary Table S2) was performed using GoTaq Green (Promega) per the manufacturer’s protocol for 40 cycles with a 60 °C annealing temperature and 1 : 30 extension time. DNA from all reported splice variants was extracted using Qiagen’s QIAquick PCR Purification Kit, and sequence was confirmed.
Immunoblots
Cells were harvested with Versene (8 g/l NaCl, 0.2 g/l KCl, 1.15 g/l Na2PO4, 0.2 g/l EDTA, 0.1 g/l Phenol red, pH 7.34), washed in ice-cold PBS and lysed on ice in a RIPA buffer with fresh protease inhibitors (Roche Complete, Roche Applied Science, Basel, Switzerland) and Phosphatase inhibitors (PhosSTOP, Roche Applied Science). Protein (25 μg) was electrophoresed through a 4–12% Bis-Tris gel, blotted to a PVDF membrane (iBLOT, Invitrogen) and immunoblotted using antibodies to: β-tubulin (Sigma T8328, Sigma-Aldrich, St. Louis, MO, USA), Mcl1 (Santa Cruz SC-819, Santa Cruz Biotechnology, Dallas, TX, USA), PSMD14 (Sigma HPA002114, Sigma-Aldrich), PRPF8 (Abcam ab79237, Cambridge, MA, USA), and SART1 (Abcam ab88583). Rabbit anti-Hub1(UBL5) antibody was a kind gift of Dr. Hideki Yashiroda (University of Tokyo, Tokyo, Japan). Blots were scanned, and densitometry was performed by the UN-SCAN-IT 6.1 software (Silk Scientific Inc, Orem, UT, USA).
Morpholino oligonucleotide transfection
IMR5 (400 000 cells/ml) and NLF (200 000 cells/ml) cells were plated in 96-well plates (cytotoxicity assays) and six-well plates (RT-PCR). Twenty-four hours after plating, the media was replaced with fresh RPMI containing 0.6% EndoPorter (GeneTools, Philomath, OR, USA) and the listed concentration of each morpholino oligonucleotide targeting the Mcl1 exon 2 splice acceptor site (5′-CGAAGCATGCCTGAGAAAGAAAAGC-3′) and splice donor site (5′-AAGGCAAACTTACCCAGCCTCTTTG-3′) or a scramble control. ABT-737 was added for cytotoxicity assays 24 h after morpholino treatment. Seventy-two hours after morpholino treatment, cytotoxicity was measured, and RNA was collected as above.
Protein half-life
IMR5 and NLF (200 000 cells/ml) cells were plated in 6-cm plates and reverse transfected with siRNA targeting PSMD14, PRPF8, UBL5 or Sham as for cytotoxicity assays. Forty-eight hours after plating, the media was replaced with fresh RPMI containing 50 μg/ml cyclohexamide (Sigma-Aldrich). After the indicated incubation period, cells were harvested for immunoblots as above.
Mutagenesis/plasmid transfection
Mutations were introduced into pcDNA-DEST40 vectors expressing full-length UBL5 (PlasmID, Harvard Medical School, Boston, MA, USA) or the PSMD14 ORF (Invitrogen) by PCR using the Invitrogen GeneArt Site Directed Mutagenesis kit per the manufacturer’s instructions. For siRNA-resistant vectors, the following mutations were introduced into the siRNA targeting region: UBL5 (Native 5′-GACCTTAAGAAGCTGATTGC-3′ to 5′-GACCTGAAGAAACTTATTGC-3′) using the primer 5′-TACCATCGGGGACCTGAAGAAACTTATTGCAGCCCAAACT-3′ and its reverse compliment, PSMD14 (Native 5′-GCATTAATTCATGGACTAAA-3′ to 5′-GCATTGATACACGGTCTAAA-3′) using the primer 5′-ATCTATCCAGGCATTGATACACGGTCTAAACAGACATTAT-3′ and its reverse compliment. The UBL5 C-terminal deletion of YYQ was introduced into the siRNA-resistant vector using the primer 5′-TGGGATGAACCTGGAGCTTTAGATGAGAATCCTCATC-3′ and its reverse compliment. The UBL5 D22A mutation (Native 5′-AACACGGATGATACCATCGGG-3′ to 5′-AACACGGATGCTACCATCGGG-3′) was introduced into the siRNA-resistant plasmid using the primer 5′-CGTTAAATGCAACACGGATGCTACCATCGGGGACCTG-3′ and its reverse compliment. The PSMD14 H113A H115A mutation (Native 5′-GGTATCACAGTCACCCTG-3′ to 5′-GGTATGCCAGTGCCCCTG-3′) was introduced into the siRNA-resistant vector using the primer 5′-GTTGTTGGTTGGTATGCCAGTGCCCCTGGCTTTGGTTG-3′ and its reverse compliment. pENTR221 expressing the Mcl1-L ORF was obtained from PlasmID. To generate Mcl1-S, exon 2 was deleted by PCR mutagenesis using the Invitrogen GeneArt Site Directed Mutagenesis kit with the primer 5′-CCACGAGACGGCCTTCCAAGGATGGGTTTGTGGAGTTCTTCC-3′ and its reverse compliment. 5′-CCTTACTGTAA-3′ was inserted after the native TAG stop codon of Mcl1-L to complete the ORF of Mcl1-S using the primer 5′-GCTGGTTTGGCATATCTAATAAGATAGCCTTACTGTAAGACCCAGCTTTCTTGTACAAAG-3′ and its reverse compliment. To express tagged Mcl1-S, the TAA stop codon of the above primer was replaced with TTA. These plasmids were cloned into the pTRex-DEST30 vector using Invitrogen LR Clonase per the manufacturer’s instructions. The insert of each vector was fully sequence confirmed. For transfection, cells were plated to be 60–80% confluent after settling overnight in a six-well plate in complete antibiotic-free media. A total of 10 μl of Lipofectamine 2000 (Invitrogen) and 4 μg of plasmid were combined in 500 μl serum-free RPMI and allowed to sit for 30 min before addition to each well. After 16-h incubation, cells were replated into a T75 flask in antibiotic-free media. Forty-eight hours after transfection, cells were selected with and maintained in RPMI supplemented with Geneticin (Invitrogen) 250 μg/ml. For tetracycline-regulated experiments, IMR5 and NLF were first transfected as above with pcDNA-TR6 (Invitrogen). Cells were selected with Blasticidin (Invitrogen) 4 μg/ml and dilutionally subcloned. Subclones were screened for expression of the TR element by western blotting using a MoBiTec antibody. Cells were then transfected with the DEST30 vector as above and selected and maintained in RPMI supplemented with Blasticidin 2 μg/ml and Geneticin 250 μg/ml.
Gene expression correlates
Publicly available NB data sets were analyzed using the R2 Microarray Analysis and Visualization platform (r2.amc.nl). The Kocak data set (GEO GSE45547) includes expression data from 649 diverse NBs with age, stage and MYCN status known, utilizing a single-color 44K oligonucleotide microarray.60 The Versteeg data set (GEO GSE16476) includes expression data from 88 NBs annotated additionally with survival outcome, utilizing the Affymetrix U133+2 genechip (Affymetrix, Santa Clara, CA, USA).61
Statistical analyses
Data are presented as mean±S.E.M. For primary screening, Z-scores were calculated as the measured ATP content minus the mean ATP content for all experimental wells divided by the S.D. For cytotoxicity curves, the viability was modeled as function of concentration and experiment group using a log-logistic regression model. All models were fitted using the ‘drm’ function in the ‘drc’ add-on package in R (R Core Team, 2013). We compared the residual sums of squares from the reduced model, which does not include experiment group variable with that from the full model, including experiment group variable to test the hypothesis of no difference between two experiments using the F-test. mRNA expression was analyzed with the two-tailed Student’s t-test using GraphPad Prism 5.0 for Macintosh (GraphPad Software, San Diego, CA, USA). Differences were considered significant at P<0.05. All data shown represent at least two independent replicates.
Abbreviations
- DUB:
-
deubiquitinase
- NB:
-
neuroblastoma
- SSA:
-
spliceostatin A
- EFS:
-
event-free survival
References
Youle RJ, Strasser A . The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol 2008; 9: 47–59.
Kim H, Rafiuddin-Shah M, Tu HC, Jeffers JR, Zambetti GP, Hsieh JJ et al. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat Cell Biol 2006; 8: 1348–1358.
Goldsmith KC, Lestini BJ, Gross M, Ip L, Bhumbla A, Zhang X et al. BH3 response profiles from neuroblastoma mitochondria predict activity of small molecule Bcl-2 family antagonists. Cell Death Differ 2010; 17: 872–882.
Goldsmith KC, Gross M, Peirce S, Luyindula D, Liu X, Vu A et al. Mitochondrial Bcl-2 family dynamics define therapy response and resistance in neuroblastoma. Cancer Res 2012; 72: 2565–2577.
Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 2005; 435: 677–681.
Lestini BJ, Goldsmith KC, Fluchel MN, Liu X, Chen NL, Goyal B et al. Mcl1 downregulation sensitizes neuroblastoma to cytotoxic chemotherapy and small molecule Bcl2-family antagonists. Cancer Biol Ther 2009; 8: 1587–1595.
Akgul C . Mcl-1 is a potential therapeutic target in multiple types of cancer. Cell Mol Life Sci 2009; 66: 1326–1336.
Bingle CD, Craig RW, Swales BM, Singleton V, Zhou P, Whyte MK . Exon skipping in Mcl-1 results in a bcl-2 homology domain 3 only gene product that promotes cell death. J Biol Chem 2000; 275: 22136–22146.
Bae J, Leo CP, Hsu SY, Hsueh AJ . MCL-1S, a splicing variant of the antiapoptotic BCL-2 family member MCL-1, encodes a proapoptotic protein possessing only the BH3 domain. J Biol Chem 2000; 275: 25255–25261.
Zhong Q, Gao W, Du F, Wang X . Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell 2005; 121: 1085–1095.
Ding Q, He X, Hsu JM, Xia W, Chen CT, Li LY et al. Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol 2007; 27: 4006–4017.
Inuzuka H, Fukushima H, Shaik S, Liu P, Lau AW, Wei W . Mcl-1 ubiquitination and destruction. Oncotarget 2011; 2: 239–244.
Vucic D, Dixit VM, Wertz IE . Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol 2011; 12: 439–452.
Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature 2009; 463: 103–107.
Ovaa H, Kessler BM, Rolen U, Galardy PJ, Ploegh HL, Masucci MG . Activity-based ubiquitin-specific protease (USP) profiling of virus-infected and malignant human cells. Proc Natl Acad Sci USA 2004; 101: 2253–2258.
McNally T, Huang Q, Janis RS, Liu Z, Olejniczak ET, Reilly RM . Structural analysis of UBL5, a novel ubiquitin-like modifier. Protein Sci 2003; 12: 1562–1566.
Beverly LJ, Lockwood WW, Shah PP, Erdjument-Bromage H, Varmus H . Ubiquitination, localization, and stability of an anti-apoptotic BCL2-like protein, BCL2L10/BCLb, are regulated by Ubiquilin1. Proc Natl Acad Sci USA 2012; 109: E119–E126.
Mishra SK, Ammon T, Popowicz GM, Krajewski M, Nagel RJ, Ares M Jr et al. Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing. Nature 2011; 474: 173–178.
Shieh JJ, Liu KT, Huang SW, Chen YJ, Hsieh TY . Modification of alternative splicing of Mcl-1 pre-mRNA using antisense morpholino oligonucleotides induces apoptosis in basal cell carcinoma cells. J Invest Dermatol 2009; 129: 2497–2506.
Apweiler R, Consortium TU . The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res 2010; 38 (Supplement 1 database issue): D142–D148.
Reed R, Hurt E . A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 2002; 108: 523–531.
Pinto AL, Steitz JA . The mammalian analogue of the yeast PRP8 splicing protein is present in the U4/5/6 small nuclear ribonucleoprotein particle and the spliceosome. Proc Natl Acad Sci USA 1989; 86: 8742–8746.
Whittaker E, Lossky M, Beggs JD . Affinity purification of spliceosomes reveals that the precursor RNA processing protein PRP8, a protein in the U5 small nuclear ribonucleoprotein particle, is a component of yeast spliceosomes. Proc Natl Acad Sci USA 1990; 87: 2216–2219.
Makarova OV, Makarov EM, Luhrmann R . The 65 and 110 kDa SR-related proteins of the U4/U6.U5 tri-snRNP are essential for the assembly of mature spliceosomes. EMBO J 2001; 20: 2553–2563.
Hegele A, Kamburov A, Grossmann A, Sourlis C, Wowro S, Weimann M et al. Dynamic protein-protein interaction wiring of the human spliceosome. Mol Cell 2012; 45: 567–580.
Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol 2007; 3: 576–583.
Berg MG, Wan L, Younis I, Diem MD, Soo M, Wang C et al. A quantitative high-throughput in vitro splicing assay identifies inhibitors of spliceosome catalysis. Mol Cell Biol 2012; 32: 1271–1283.
Versteeg R R2: microarray analysis and visualization platform (http://r2.amc.nl).
Hanahan D, Weinberg RA . Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674.
Davids MS, Deng J, Wiestner A, Lannutti BJ, Wang L, Wu CJ et al. Decreased mitochondrial apoptotic priming underlies stroma-mediated treatment resistance in chronic lymphocytic leukemia. Blood 2012; 120: 3501–3509.
Roberts AW, Seymour JF, Brown JR, Wierda WG, Kipps TJ, Khaw SL et al. Substantial susceptibility of chronic lymphocytic leukemia to BCL2 inhibition: results of a phase I study of navitoclax in patients with relapsed or refractory disease. J Clin Oncol 2012; 30: 488–496.
van Delft MF, Wei AH, Mason KD, Vandenberg CJ, Chen L, Czabotar PE et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell 2006; 10: 389–399.
Morales AA, Kurtoglu M, Matulis SM, Liu J, Siefker D, Gutman DM et al. Distribution of Bim determines Mcl-1 dependence or codependence with Bcl-xL/Bcl-2 in Mcl-1-expressing myeloma cells. Blood 2011; 118: 1329–1339.
D'Arcy P, Brnjic S, Olofsson MH, Fryknas M, Lindsten K, De Cesare M et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat Med 2011; 17: 1636–1640.
Sveda M, Castoralova M, Lipov J, Ruml T, Knejzlik Z . Human UBL5 protein interacts with coilin and meets the Cajal bodies. Biochem Biophys Res Commun 2013; 436: 240–245.
van der Feltz C, Anthony K, Brilot A, Pomeranz Krummel DA . Architecture of the spliceosome. Biochemistry (Mosc) 2012; 51: 3321–3333.
Roybal GA, Jurica MS . Spliceostatin A inhibits spliceosome assembly subsequent to prespliceosome formation. Nucleic Acids Res 2010; 38: 6664–6672.
Moore MJ, Wang Q, Kennedy CJ, Silver PA . An alternative splicing network links cell-cycle control to apoptosis. Cell 2010; 142: 625–636.
Gautrey HL, Tyson-Capper AJ . Regulation of Mcl-1 by SRSF1 and SRSF5 in cancer cells. PLoS One 2012; 7: e51497.
Michlewski G, Sanford JR, Caceres JF . The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell 2008; 30: 179–189.
Mills JR, Hippo Y, Robert F, Chen SM, Malina A, Lin CJ et al. mTORC1 promotes survival through translational control of Mcl-1. Proc Natl Acad Sci USA 2008; 105: 10853–10858.
Schwerk C, Schulze-Osthoff K . Regulation of apoptosis by alternative pre-mRNA splicing. Mol Cell 2005; 19: 1–13.
Akgul C, Moulding DA, Edwards SW . Alternative splicing of Bcl-2-related genes: functional consequences and potential therapeutic applications. Cell Mol Life Sci 2004; 61: 2189–2199.
Grabowski P . Alternative splicing takes shape during neuronal development. Curr Opin Genet Dev 2011; 21: 388–394.
Lim DA, Suarez-Farinas M, Naef F, Hacker CR, Menn B, Takebayashi H et al. In vivo transcriptional profile analysis reveals RNA splicing and chromatin remodeling as prominent processes for adult neurogenesis. Mol Cell Neurosci 2006; 31: 131–148.
An M, Henion PD . The zebrafish sf3b1b460 mutant reveals differential requirements for the sf3b1 pre-mRNA processing gene during neural crest development. Int J Dev Biol 2012; 56: 223–237.
Keightley MC, Crowhurst MO, Layton JE, Beilharz T, Markmiller S, Varma S et al. In vivo mutation of pre-mRNA processing factor 8 (Prpf8) affects transcript splicing, cell survival and myeloid differentiation. FEBS Lett 2013; 587: 2150–2157.
Pena V, Liu S, Bujnicki JM, Luhrmann R, Wahl MC . Structure of a multipartite protein-protein interaction domain in splicing factor prp8 and its link to retinitis pigmentosa. Mol Cell 2007; 25: 615–624.
Zhang B, Gojo I, Fenton RG . Myeloid cell factor-1 is a critical survival factor for multiple myeloma. Blood 2002; 99: 1885–1893.
Tiedemann RE, Zhu YX, Schmidt J, Shi CX, Sereduk C, Yin H et al. Identification of molecular vulnerabilities in human multiple myeloma cells by RNA interference lethality screening of the druggable genome. Cancer Res 2012; 72: 757–768.
Bonnal S, Vigevani L, Valcarcel J . The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov 2012; 11: 847–859.
Maciejewski JP, Padgett RA . Defects in spliceosomal machinery: a new pathway of leukaemogenesis. Br J Haematol 2012; 158: 165–173.
Bergen HR III, Vasmatzis G, Cliby WA, Johnson KL, Oberg AL, Muddiman DC . Discovery of ovarian cancer biomarkers in serum using NanoLC electrospray ionization TOF and FT-ICR mass spectrometry. Dis Markers 2003; 19: 239–249.
van Alphen RJ, Wiemer EA, Burger H, Eskens FA . The spliceosome as target for anticancer treatment. Br J Cancer 2009; 100: 228–232.
Nakajima H, Hori Y, Terano H, Okuhara M, Manda T, Matsumoto S et al. New antitumor substances, FR901463, FR901464 and FR901465. II. Activities against experimental tumors in mice and mechanism of action. J Antibiot 1996; 49: 1204–1211.
Lagisetti C, Pourpak A, Goronga T, Jiang Q, Cui X, Hyle J et al. Synthetic mRNA splicing modulator compounds with in vivo antitumor activity. J Med Chem 2009; 52: 6979–6990.
Gao Y, Koide K . Chemical perturbation of Mcl-1 pre-mRNA splicing to induce apoptosis in cancer cells. ACS Chem Biol 2013; 8: 895–900.
Tumilowicz JJ, Nichols WW, Cholon JJ, Greene AE . Definition of a continuous human cell line derived from neuroblastoma. Cancer Res 1970; 30: 2110–2118.
Schlesinger HR, Gerson JM, Moorhead PS, Maguire H, Hummeler K . Establishment and characterization of human neuroblastoma cell lines. Cancer Res 1976; 36 (9 pt.1): 3094–3100.
Kocak H, Ackermann S, Hero B, Kahlert Y, Oberthuer A, Juraeva D et al. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell Death Dis 2013; 4: e586.
Molenaar JJ, Domingo-Fernandez R, Ebus ME, Lindner S, Koster J, Drabek K et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat Genetics 2012; 44: 1199–1206.
Acknowledgements
We thank Dr. Minoru Yoshisa (RIKEN Institute, Japan) for the gift of Spliceostatin A, Dr. Gideon Dreyfuss (University of Pennsylvania) for BN82685, Dr. Kazunori Koide (University of Pittsburgh) for Meayamycin B, Dr. Jill Lahti and Dr. Thomas Webb (St. Jude Children’s Research Center) for Sudemycin C1 and E, and Dr. Koji Ikegami (Hamamatsu University School of Medicine, Japan) and Dr. Hideki Yashiroda (University of Tokyo) for rabbit anti-Hub1 antibody. We acknowledge the helpful advice of Serge Fuchs (University of Pennsylvania) concerning DUB protease activities and Kristen Lynch (University of Pennsylvania) concerning spliceosome activities. This work was supported by NIH CA97323, Alex’s Lemonade Stand Foundation, Cookies for Kids Cancer Foundation (to MDH) and Bear Necessities Pediatric Cancer Foundation and NIH T32-HD0433021 (to TWL).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Raschellá
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Laetsch, T., Liu, X., Vu, A. et al. Multiple components of the spliceosome regulate Mcl1 activity in neuroblastoma. Cell Death Dis 5, e1072 (2014). https://doi.org/10.1038/cddis.2014.40
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2014.40