Abstract
Many mitotic kinases are both critical for maintaining genome stability and are important targets for anticancer therapies. We provide evidence that SIK3 (salt-inducible kinase 3), an AMP-activated protein kinase-related kinase, is important for mitosis to occur properly in mammalian cells. Downregulation of SIK3 resulted in an extension of mitosis in both mouse and human cells but did not affect the DNA damage checkpoint. Time-lapse microscopy and other approaches indicated that mitotic exit but not mitotic entry was delayed. Although repression of SIK3 alone simply delayed mitotic exit, it was able to sensitize cells to various antimitotic chemicals. Both mitotic arrest and cell death caused by spindle poisons were enhanced after SIK3 depletion. Likewise, the antimitotic effects due to pharmacological inhibition of mitotic kinases including Aurora A, Aurora B, and polo-like kinase 1 were enhanced in the absence of SIK3. Finally, in addition to promoting the sensitivity of a small-molecule inhibitor of the mitotic kinesin Eg5, SIK3 depletion was able to overcome cells that developed drug resistance. These results establish the importance of SIK3 as a mitotic regulator and underscore the potential of SIK3 as a druggable antimitotic target.
Similar content being viewed by others
Main
Mitosis is associated with a profound surge in protein phosphorylation. A remarkably large portion of the kinome is involved to ensure the timely and proper execution of mitosis, including archetypal mitotic kinases such as cyclin-dependent kinase, Aurora kinases, NIMA-like kinases, and polo-like kinases (PLKs).1 By depleting all Drosophila kinases in cultured S2 cells, Bettencourt-Dias et al.2 found that downregulation of a large subset of the kinome (60 out of 228 kinases) results in mitotic dysfunction. In addition to established mitotic kinases such as PLK, many candidates were unexpected either because they had not been previously characterized or were known to have other functions. The mammalian protein kinase complement is about double the size of Drosophila’s.3 Whether a similar set of mammalian protein kinases are involved in mitotic control as in Drosophila remains to be deciphered.
SIK1 (salt-inducible kinase 1) (also called SIK or SNF1LK) was isolated from adrenal glands of high-salt diet-fed rats.4 Together with two other isoforms, SIK2 (also called QIK or SNF1LK2) and SIK3 (also called QSK), they belong to a subfamily of serine/threonine protein kinase with similarity to the kinase domain of the AMP-activated protein kinase (AMPK) family. Similar to most AMPK-related proteins, the T-loop of SIK3 can be phosphorylated in vitro by LKB1.5 This phosphorylation generates a 14-3-3 binding site, which promotes the catalytic activity and localization of SIK3 to punctate structures within the cytoplasm.6
Several functions have been implicated for SIK3, including energy balance and growth control. SIK3 phosphorylates class IIa histone deacetylases (HDACs), thereby stimulating 14-3-3 binding and nucleocytoplasmic trafficking.7 SIK3 is also inactivated during fasting in Drosophila, leading to the dephosphorylation and nuclear accumulation of HDAC4. The HDAC4 then deacetylates the forkhead factor FOXO, allowing the shift from glucose to fat burning to maintain energy balance.8 Accordingly, SIK3-deficient mice is characterized by defects in glucose and lipid homeostasis.9 The mislocalization of HDAC4 may also explain the severe inhibition of chondrocyte hypertrophy during skeletal development in SIK3-deficient mice.10 In Drosophila, SIK3 is also a negative regulator the Hippo pathway, a major mechanism for promoting cell cycle exit and apoptosis.11 SIK3 was found to be overexpressed in a subset of ovarian cancer.12 Moreover, overexpression of SIK3 in ovarian cancer cells stimulates cell growth by attenuating the expression of the p21CIP1/WAF1 and p27KIP1.12
Deregulation of many of the mitotic kinases contributes to genome instability and cancer. On the other hand, targeting several mitotic kinases can enhance the effectiveness of antimitotic drugs in clinical use. In this study, using a kinome-wide screen for siRNAs that extended mitotic length in mouse fibroblasts, we identified a number of novel kinases involved in mitotic progression. We further demonstrated that SIK3 is important for mitotic exit in human and mouse cells. Depletion of SIK3 could increase the sensitivity of cancer cells to several antimitotic drugs, including inhibitors microtubules, kinesin, and several mitotic kinases.
Results
Kinome-wide screen identifies Sik3 to be important for mitosis in mouse fibroblasts
An NIH3T3 mouse fibroblast cell line expressing GFP-tagged histone H3 was generated to facilitate the imaging of mitosis in living cells. The NIH3T3/H2B-GFP cells were transfected with an siRNA library targeting the mouse kinome complement (also included some phosphotransferases and protein kinase regulators). Each of the 588 genes in the library was targeted by three siRNAs (Supplementary Figure S1). We used a strategy that involved addition of a DNA-damaging agent (Adriamycin) at 2.5 h before analysis. Activation of the G2 DNA damage checkpoint prevented entry into mitosis but not the progression of cells already in mitosis. Therefore, while the mitotic population in siRNA transfection not affecting mitosis was effectively reduced to nil, it remained elevated for siRNAs that resulted in a delay in mitotic progression. Using this approach, 20 candidates were found to increase the mitotic index above 2 × S.D. of the mean (Figure 1a). Although the screen should also reveal kinases important for the DNA damage checkpoint, we found that most of the candidate siRNAs increased the mitotic index even in the absence of DNA damage (Figure 1b), suggesting that these kinases were involved in regulating mitosis instead of the DNA damage checkpoint.
Identification of kinases important for mitosis in mouse fibroblasts. (a) Screening of the mouse kinome for mitotic kinases. NIH3T3/H2B-GFP cells were transfected with a small interfering RNA (siRNA) library targeting the mouse kinome and related genes. Each of the 588 genes in the library was targeted by three different Stealth siRNAs. After 24 h, the cells were treated with 0.2 μg/ml of Adriamycin for 2.5 h. Images of the cells were then captured automatically using a fluorescent microscopy and the mitotic index of each transfection was scored. Cutoff was set to candidates with mitotic index above 2 × S.D. of the mean. The mitotic index of these candidate genes are shown. Note that non-protein kinases are also included in the library: Dgkg (diacylglycerol kinase gamma); Mpp2 (MAGUK p55 subfamily member 2 guanylate kinase); Prps1 (ribose-phosphate pyrophosphokinase 1); Sphk2 (sphingosine kinase 2). (b) The candidates obtained from the initial screen (a) were subjected to secondary confirmation assays in the absence of Adriamycin. NIH3T3/H2B-GFP cells were transfected with siRNAs against the indicated candidates. Each gene was targeted by three different Stealth siRNAs. After 26.5 h, the mitotic index was quantified using a fluorescent microscopy. The dotted line indicates the mitotic index of the control. (c) Comparison between mouse and Drosophila kinome required for mitosis. The protein kinases found to be important for mitosis in mouse cells in this study are compared with protein kinases found to contribute to mitosis in Drosophila.2 Only four homologous candidates are common in the two screens
Intriguingly, the protein kinases identified in our screen has only limited overlap with those identified in a pioneering kinome RNAi screen using Drosophila S2 cells.2 Four protein kinases were found to be common targets for mitotic regulation in both mouse and Drosophila cells (Figure 1c). As expected, the PLK1 (polo in Drosophila) was a potent mitotic regulator in both mouse and Drosophila models. Other common candidates include Sik3 (CG15072), Scyl1 (CG1951), and Tbk1 (ik2) (Drosophila proteins are indicated in the brackets). Given that Sik3 was found to be important for mitosis in both mouse and Drosophila cells, and that other AMPK-related kinases (such as Brsk2; Figure 1a) were also identified in both screens, we further characterized the role of Sik3 in mitosis in this study.
Depletion of SIK3 increases the duration of mitosis
SIK3 (salt-inducible kinase 3, also called QSK) is a member of AMPK family. As the original screen involved the use of a mixture of siRNAs against each kinase, we first verified the results for Sik3 using the different siRNAs individually. Figure 2a shows that transfection with the three Sik3 siRNAs increased the mitotic index in NIH3T3 fibroblasts irrespective of the presence or absence of Adriamycin-mediated DNA damage, supporting the specificity of the mitotic effects for Sik3.
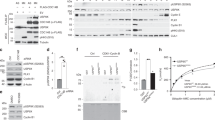
Depletion of SIK3 increases the mitotic population in mouse and human cell lines. (a) Transfection of Sik3 small interfering RNA (siRNA) increases the mitotic index in mouse fibroblasts. NIH3T3/H2B-GFP cells were transfected with control siRNA or three independent siRNAs against Sik3 (siSik3). After 24 h, the cells were treated with either buffer (lower panel) or 0.2 μg/ml of Adriamycin (upper panel) for 2.5 h. The mitotic index was then quantified using fluorescent microscopy (average±S.D. from three independent experiments). At least two out of the three siSik3 used in the original screen were able to increase the mitotic index significantly (P<0.05; unpaired t-test). (b) SIK3 is ubiquitously expressed in human cells from different origins. Lysates of various cell lines were prepared and analyzed with immunoblotting for SIK3. Cells from normal fibroblasts (IMR90), immortalized normal nasopharyngeal epithelial (NP460, NP361, and NP550), nasopharyngeal carcinoma (CNE2, HNE1, HONE1, and C666-1), immortalized normal liver epithelial (LO2 and MIHA), and liver cancer (Hep3B and HepG2) were analyzed. Same amount of total protein was loaded for different cell lines. (c) Depletion of SIK3 in human cells. HeLa cells were transfected with control siRNA or three independent siRNAs against SIK3 (siSIK3). The cells were harvested after 24 h. Lysates were prepared and subjected to immunoprecipitation (IP) with an SIK3 antiserum. Both the total lysates and immunoprecipitates were analyzed with immunoblotting with antibodies against SIK3. Actin analysis was included to assess protein loading and transfer. (d) Depletion of SIK3 increases the mitotic index in HeLa cells. HeLa/H2B-GFP cells were transfected with control siRNA or siSIK3. Transfection with siPLK1 served as a positive control. After 24 h, the mitotic index was quantified (average±S.D. from three independent experiments). Depletion of SIK3 significantly increased the mitotic index (P<0.05; unpaired t-test). (e) Depletion of SIK3 increases the mitotic index in H1299 cells. H1299/H2B-GFP cells were transfected with control siRNA, siPLK1, or siSIK3. After 24 h, the mitotic index was quantified (average±S.D. from three independent experiments). Depletion of SIK3 significantly increased the mitotic index (P<0.05; unpaired t-test). (f) Depletion of SIK3 increases population containing phosphorylated histone H3Ser10. HeLa cells were transfected with either control siRNA or siSIK3. After 24 h, the cells were fixed and analyzed with bivariate flow cytometry analysis (DNA content and phosphorylated histone H3Ser10). Histone H3Ser10 phosphorylation-positive cells (arrows) were quantified (lower panel; average±S.D. from three independent experiments)
We next investigated if the mitotic effects of SIK3 are conserved in human cells. Figure 2b shows that SIK3 was expressed in most cell lines we examined, including several relatively normal (fibroblasts and immortalized normal epithelial cells from liver and nasopharynx) and cancer cell lines (from liver and nasopharynx, except C666-1). To evaluate if the expression of SIK3 affects mitosis in human cells, HeLa cells were transfected with three siRNAs targeting different regions of human SIK3. All three siRNAs were able to downregulate SIK3 protein effectively (Figure 2c). While the results from siSIK3 no. 1 were mainly used in this paper, similar results were obtained with at least one other siSIK3s. As in mouse fibroblasts, knockdown of SIK3 in HeLa cells increased the mitotic index by 1.5–2-fold (Figure 2d). Similar results were obtained using H1299 cells (Figure 2e), indicating that the effect was not only specific for HeLa cells. Although the increase in mitotic index was less impressive than after depletion of the classic mitotic kinase PLK1 (Figures 2d and e), it was nevertheless significant and reproducible.
We further analyzed the mitotic population using bivariate flow cytometry to measure DNA contents and histone H3Ser10 phosphorylation together. Figure 2f shows that more siSIK3-transfected cells displayed phosphorylated histone H3Ser10 than in control cells, verifying that depletion of SIK3 increased the mitotic population.
To demonstrate more directly the increase in mitotic duration, HeLa cells expressing histone H2B-GFP were transfected with siSIK3 and followed at single-cell levels with time-lapse microscopy. The average duration of mitosis was increased following siSIK3 transfection (Figure 3a) (the complete fates of individual cells are shown in Supplementary Figure S2A). The increase in mitotic duration was reproducible with two siSIK3s (Figure 3b), both of which were able to reduce the expression of endogenous SIK3 (Figure 3c). Finally, siSIK3 also extended mitosis in another cell line (Hep3B), excluding the possibility that the effect was only specific for HeLa cells (Figure 3d).
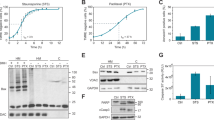
Depletion of SIK3 increases the duration of mitosis in mouse and human cell lines. (a) Depletion of SIK3 increases the duration of mitosis in HeLa cells. HeLa/H2B-GFP cells were transfected with control small interfering RNA (siRNA), siSIK3, or siPLK1. After 24 h, the cells were analyzed with time-lapse microscopy. Individual cells were tracked for 24 h to measure the duration of mitosis (from DNA condensation to anaphase or cell death). Each horizontal bar represents the duration of mitosis for one cell (n=50). (b) siSIK3 increases the duration of mitosis in HeLa cells. HeLa/H2B-GFP cells were transfected with control siRNA or two different siSIK3. After 24 h, the cells were analyzed with time-lapse microscopy. The duration of mitosis was quantified (average±95% CI). Unpaired t-test: P<0.05 and 0.01 for siSIK3 no. 1 and siSIK3 no. 2, respectively. (c) Downregulation of SIK3 with siRNAs. HeLa/H2B-GFP cells were transfected with control siRNA or two different siSIK3. The cells were harvested after 24 h and analyzed with immunoblotting. FLAG-HA-tagged SIK3 expressed in HeLa cells was used as a standard. (d) siSIK3 increases the duration of mitosis in Hep3B cells. Hep3B/H2B-GFP cells were transfected with control siRNA or siSIK3. After 24 h, the cells were analyzed with time-lapse microscopy. The duration of mitosis was quantified (average±95% CI). (e) siSIK3 delays mitotic exit. HeLa/H2B-GFP cells were transfected with control siRNA or siSIK3. After 24 h, the cells were analyzed with time-lapse microscopy. Representative control siRNA- and siSIK3-transfected cells are shown
As SIK3 was originally identified in our screen in the presence of DNA damage (Figure 1), we also verified that siSIK3 did not abrogate the DNA damage checkpoint in human cells. Exposing HeLa cells to ionizing radiation abolished mitotic entry (Supplementary Figure S2B). While incubation with a CHK1 inhibitor (UCN-01) overrode the checkpoint and triggered precocious mitotic entry as expected, depletion of SIK3 did not affect the cell cycle arrest. Collectively, these data show that downregulation of SIK3 resulted in an increase in mitotic duration in both mouse and human cells but did not affect the DNA damage checkpoint.
SIK3 is required for proper mitotic exit
More detailed analysis revealed that metaphase plate could form normally after SIK3 depletion. However, onset of anaphase was significantly delayed. The metaphase plate typically flipped and rotated for an extensive period of time before anaphase (Figure 3e). Another example is shown in Supplementary Video S1.
To examine the mitotic defects in more synchronized population, cells were trapped at prometaphase with nocodazole after they were first released from a double thymidine block. Flow cytometry revealed that after they were released from the nocodazole-mediated block, control cells were in G1 at t=2 h. In contrast, entry into G1 was delayed in siSIK3-expressing cells (Figure 4a). In agreement with this, degradation of several anaphase-promoting complex/cyclosome (APC/C) targets, including cyclin B1 and securin, as well as dephosphorylation of histone H3Ser10, was delayed in siSIK3-transfected cells (Figure 4b). Finer temporal resolution using time-lapse microscopy confirmed the delay of anaphase onset (Figure 4c). To determine if mitotic entry is also affected by siSIK3, cells were synchronized at the G2 phase with the CDK1 inhibitor RO3306 and released into mitosis. We found that while the timing of entry into mitosis was not affected, anaphase onset was delayed in the absence of SIK3 (Figure 4d). Taken together, these results indicate that mitotic exit but not mitotic entry was delayed in the absence of SIK3.
Depletion of SIK3 induces defective mitotic exit. (a) SIK3 depletion delays mitotic exit. HeLa cells expressing histone H2B-GFP were transfected with control small interfering RNA (siRNA) or siSIK3. The cells then synchronized at prometaphase by blocking cells with nocodazole after they were released from a double thymidine procedure. After releasing from the nocodazole-mediated block, the cells were harvested at the indicated time points and analyzed with flow cytometry. The percentage of G2/M cells was quantified and plotted on the right (average±S.D. from three independent experiments). Mitotic exit was significantly slower in siSIK3-treated cells (P<0.01 at 2 h after release; unpaired t-test). (b) SIK3 depletion delays degradation of APC/C targets. HeLa/H2B-GFP cells were transfected and synchronized with nocodazole as described in a. Cell-free extracts were prepared and the expression of the indicated proteins was detected with immunoblotting. Equal loading of lysates was confirmed by immunoblotting for actin. The abundance of cyclin B1 was quantified with densitometry (average±S.D. from three independent experiments). Degradation of cyclin B1 was significantly slower in siSIK3-treated cells (P<0.05 at 2and 3 h after release; unpaired t-test). (c) SIK3 depletion delays mitotic exit. HeLa/H2B-GFP cells were transfected and synchronized with nocodazole as described in a. Individual cells were tracked with time-lapse microscopy to measure the time of anaphase onset. Transfection of siSIK3 significantly delayed anaphase onset (P=0.0087; log-rank (Mantel–Cox) test). (d) SIK3 depletion does not affect mitotic entry. HeLa/H2B-GFP cells were transfected with control siRNA or siSIK3. The cells were then synchronized at the G2 phase with the CDK1 inhibitor RO3306 and released into mitosis. Individual cells were tracked with time-lapse microscopy to measure the time of entry into mitosis and anaphase onset. Transfection of siSIK3 did not affect the time of mitotic entry (P>0.1) but significantly delayed anaphase onset (P<0.0001; log-rank (Mantel–Cox) test)
SIK3 expression modulates the sensitivity to spindle poisons
Given that depletion of SIK3 results in aberrant mitotic exit, we next investigated if the efficacy of antimitotic chemicals can be altered by SIK3. Classic spindle poisons such as nocodazole (an inhibitor of microtubule polymerization) activate the spindle-assembly checkpoint and trap cells in mitosis. A relatively low concentration of nocodazole was used in these experiments with the aim of delaying mitosis without inducing extensive mitotic cell death. As expected, mitosis was extended after treatment with siSIK3 or nocodazole individually (Figure 5a). When siSIK3 and nocodazole were applied together, the cells were trapped in mitosis for an extensive period of time before undergoing apoptosis. Similar to siSIK3 alone, the metaphase–anaphase transition was markedly delayed when siSIK3 and nocodazole were added together (Figure 5b and Supplementary Video S2).
Downregulation of SIK3 increases the cytotoxicity of spindle poisons. (a) SIK3 depletion increases nocodazole-mediated mitotic cell death in HeLa cells. HeLa cells expressing histone H2B-GFP were transfected with control small interfering RNA (siRNA) or siSIK3. After 24 h, the cells were treated with 6.25 ng/ml of nocodazole. After 2 h, individual cells were tracked using time-lapse microscopy for 24 h. Each horizontal bar represents one cell (n=50). Key: gray=interphase; black=mitosis (from DNA condensation to anaphase or cell death); truncated bars=cell death. (b) The duration of mitosis after nocodazole treatment is increased after SIK3 depletion. HeLa/H2B-GFP cells were transfected, challenged with nocodazole, and imaged with time-lapse microscopy as described in a. The duration from prometaphase to metaphase and from metaphase to anaphase was quantified (average±90% CI). Transfection of siSIK3 significantly increased the duration of mitosis after nocodazole challenge (P<0.0001; unpaired t-test)
The stimulation of spindle poison-mediated cell death by siSIK3 was not limited to HeLa cells, as similar results were found with the colon carcinoma HCT116 cells (Supplementary Figure S3). Depletion of SIK3 also enhanced mitotic cell death following challenge with Taxol (an inhibitor of microtubule depolymerization), indicating that siSIK3 enhanced the effects of multiple spindle poisons (Supplementary Figure S3). Taken together, these results suggest that downregulation of SIK3 can sensitize cells to mitotic arrest and cell death induced by spindle poisons.
SIK3 expression affects the sensitivity to pharmacological inhibition of multiple mitotic kinases
The shortcomings of spindle poisons as chemotherapeutic agents include neuropathy and drug resistance. Newer generations of antimitotic drugs are developed to target specific mitotic components such as Aurora kinases. Aurora A (AURKA) is a centrosomal protein that regulates the maturation and separation of centrosomes and formation of bipolar spindle; Aurora B (AURKB) is a component of the chromosomal passenger complex that mainly regulates late mitotic events.1 We used a small-molecule inhibitor Alisertib (also called MLN8237) that inhibits both AURKA and AURKB,13, 14 as well as newer generations of specific inhibitors of AURKA (MK-5108, also called VX-689)15 and AURKB (Barasertib, also called AZD1152-HQPA).16 As complete pharmacological inhibition of AURKA and AURKB triggers an early mitotic arrest and mitotic slippage, respectively,14 cells were treated with relatively low concentrations of the drugs that were insufficient to trigger these effects on their own. Figure 6a shows that depletion of SIK3 significantly enhanced the G2/M delay induced by these Aurora inhibitors.
Downregulation of SIK3 increases the sensitivity to inhibitors of Aurora kinases and PLK1. (a) SIK3 depletion increases the sensitivity to inhibitors of Aurora kinases and PLK1. HeLa cells were transfected with control small interfering RNA (siRNA) or siSIK3. After 24 h, the cells were treated with buffer, AURKAi (MK-5108), AURKBi (Barasertib), AURKA/Bi (Alisertib), or PLK1i (BI-2536), as described in Materials and Methods. After another 24 h, the cells were harvested and analyzed with flow cytometry. (b) SIK3 depletion promotes AURKBi-induced mitotic slippage. HeLa cells expressing histone H2B-GFP were transfected with control siRNA or siSIK3. After 24 h, the cells were treated with 12.5 nM of Barasertib. After 2 h, individual cells were tracked using time-lapse microscopy for 24 h. Each horizontal bar represents one cell (n=50). Key: light gray=interphase; black=mitosis (from DNA condensation to anaphase or mitotic slippage); dark gray=interphase after mitotic slippage; truncated bars=cell death. (c) SIK3 depletion promotes PLK1i-mediated mitotic cell death. HeLa cells expressing histone H2B-GFP were transfected with control siRNA or siSIK3. After 24 h, the cells were treated with 1.25 nM of BI-2536. After 2 h, individual cells were tracked using time-lapse microscopy for 24 h. Each horizontal bar represents one cell (n=50). Key: light gray=interphase; black=mitosis (from DNA condensation to anaphase or mitotic slippage); truncated bars=cell death. (d) The onset of anaphase is delayed after inhibition of PLK1 and depletion of SIK3. HeLa/H2B-GFP cells were transfected, challenged with BI-2536, and imaged with time-lapse microscopy as described in c. The duration from prometaphase to metaphase and from metaphase to anaphase was quantified (average±90% CI). Transfection of siSIK3 significantly increased the duration of mitosis after BI-2536 challenge (P<0.05; unpaired t-test)
The main effect of AURKB inhibition is mitotic slippage, encompassing of premature mitotic exit without sister chromatid separation.14 Live-cell imaging indicated that the increase in G2/M population after codepletion of SIK3 was caused by an exacerbation of mitotic slippage (Figure 6b and Supplementary Video S3).
In contrast to inhibition of Aurora kinases, inhibition of the PLK1 induces a metaphase arrest. We found that siSIK3 collaborated with the PLK1 inhibitor BI-253617 in promoting G2/M delay (Figure 6a). Live-cell imaging indicated that the cells were trapped in mitosis before undergoing apoptosis (Figure 6c). As both siSIK3 and BI-2536 individually delayed metaphase–anaphase transition, it is not surprising that cells treated with both reagents were arrested at metaphase (Figure 6d and Supplementary Video S4). Collectively, these data indicate that the mitotic defects and cytotoxicity induced by small-molecule inhibitors of AURKA, AURKB, and PLK1 are substantially enhanced in the absence of SIK3.
Antimitotic drug resistance can be overcome by SIK3 depletion
Eg5 (also called kinesin family member 11, KIF11) is a plus-end-directed microtubule motor of the kinesin-5 family. Eg5 is implicated in various mitotic microtubule functions, including microtubule crosslinking, antiparallel microtubule sliding, and bipolar spindle formation, ensuring the fidelity of chromosome segregation. We found that the effects of pharmacological inhibition of Eg5 could be enhanced in the absence of SIK3. While depletion of SIK3 or a relatively low concentration of the Eg5 inhibitor SB74392118 alone did not affect cell cycle distribution, treatment of siSIK3 and SB743921 together triggered a significant G2/M delay (Figure 7a). Live-cell imaging revealed that the cells were arrested in mitosis before undergoing apoptosis (Figure 7b).
Downregulation of SIK3 increases the sensitivity to antimitotic drugs. (a) SIK3 depletion enhances the sensitivity to the Eg5 inhibitor (Eg5i) SB743921. HeLa cells were transfected with control small interfering RNA (siRNA) or siSIK3. After 24 h, the cells were treated with buffer or 1.25 nM of SB743921. After another 24 h, the cells were harvested, fixed, and analyzed with flow cytometry. (b) SIK3 depletion promotes SB743921-mediated mitotic arrest. HeLa cells expressing histone H2B-GFP were transfected with control siRNA or siSIK3. After 24 h, the cells were treated with 1.25 nM of SB743921. After 2 h, individual cells were tracked using time-lapse microscopy for 24 h. Each horizontal bar represents one cell (n=50). Key: light gray=interphase; black=mitosis (from DNA condensation to anaphase or mitotic slippage); truncated bars=cell death. (c) Early mitotic delay after inhibition of Eg5 and depletion of SIK3. HeLa/H2B-GFP cells were transfected, challenged with SB743921, and imaged with time-lapse microscopy as described in b. The duration from prometaphase to metaphase and from metaphase to anaphase was quantified (average±90% CI). Transfection of siSIK3 significantly increased prometaphase delay after SB743921 challenge (P<0.0001; unpaired t-test). (d) SIK3 depletion enhances the sensitivity to SB743921 in drug-resistant cells. HeLa cells and two independent clones of SB743921-resistant cells (SBR no. 1 and SBR no. 4) were transfected with control siRNA or siSIK3. After 24 h, the cells were treated with buffer or 10 nM of SB743921. After another 24 h, the cells were harvested, fixed, and analyzed with flow cytometry. (e) SIK3 depletion does not enhance the mitotic defects induced by the HDAC inhibitor SAHA. HeLa cells expressing histone H2B-GFP were transfected with control siRNA or siSIK3. After 24 h, the cells were treated with 1 nM of SAHA. After 2 h, individual cells were tracked using time-lapse microscopy for 24 h. The duration from prometaphase to metaphase and from metaphase to anaphase was quantified (n=50, average±90% CI). There is no significant additive effects of the two treatments (P>0.1; unpaired t-test)
Notably, inhibition of Eg5 or SIK3 produced rather different effects on mitosis. While SB743921 delayed early mitosis with monopolar spindle, siSIK3 delayed mitosis mainly after the formation of metaphase plate. Interestingly, when both SB743921 and siSIK3 were added together, the cells were trapped in early mitosis without forming the metaphase plate (Figure 7c and Supplementary Video S5). This suggested that the effect of Eg5 inhibition was exacerbated in the absence of SIK3.
Development of drug resistance is one of the major obstacles of anticancer therapies. To generate SB743921-resistant cells, we propagated HeLa cells in medium containing progressive increasing concentrations of SB743921. Figure 7d shows that while the parental HeLa cells could be arrested at mitosis with 10 nM of SB743921, the SB743921-resistant cells were unaffected by the same concentration of SB743921 (two independent clones are shown). Significantly, after SIK3 was depleted with siSIK3, both drug-resistant clones could be arrested at mitosis with SB743921.
Taken together, these results suggest that depletion of SIK3 can potentiate the effects of several antimitotic drugs on cancer cells as well as drug-resistant clones, implying that SIK3 could be a potential target for anticancer therapies.
SIK3 may exert mitotic functions through HDACs
As only a few substrates of SIK3 are known, the molecular basis of how SIK3 regulates mitosis remains to be defined. As class IIa HDACs can be phosphorylated by SIK3,7 one possibility is that SIK3 exerts its effects on mitosis indirectly through HDACs. For example, downregulation of HDAC4 with RNAi in HeLa cells was shown to induce mitotic arrest;19 and small chemical inhibitors of HDACs were found to cause a variety of chromosome segregation defects.20
In this connection, we have tested if SIK3 depletion enhances the mitotic defects of HDAC inhibition. SAHA (also called Vorinostat or Zolinza or MK0683) is an FDA-approved pan-HDAC inhibitor for cutaneous T-cell lymphoma.21 Incubation of HeLa cells with 1 nM of SAHA resulted in a delay of mitotic exit. However, depletion of SIK3 did not further delay mitosis induced by SAHA (Figure 7e), indicating that depletion of SIK3 did not act synergistically with all antimitotic drugs. These data are also consistent with the model that SIK3 may function in the same pathway as HDAC in disrupting mitosis.
Discussion
SIK3 as a novel mitotic regulator and target for antimitotic drugs
Using a simple kinome-wide RNAi screen, we found that downregulation of Sik3 increased the mitotic population in mouse fibroblasts (Figure 1). We characterized Sik3 in particular because downregulation of the Drosophila homolog (CG15072) was also reported to affect mitosis, in particular on spindle morphology,2 suggesting a possible conservation of mitotic functions for this kinase. Another determining factor is that downregulation of several AMPK-related proteins, including SNF1A in Drosophila2 and BRSK2 in mouse (Figure 1b) and human cells (data not shown), also affected mitosis.
Interestingly, downregulation of another AMPK-related protein BRSK2 also increased the mitotic duration in mouse and human cells (Figure 1). Given that both SIK3 and BRSK2 are downstream targets of LKB1,5 it is possible that LKB1-deficient cells are also sensitized to antimitotic drugs. However, the effect will probably be complicated by the fact that LKB1 is a master regulator of 13 AMPK-related kinases.
We first confirmed that SIK3 could be effectively downregulated with siRNAs in human cells (Figure 2c). Several methods including mitotic index count (Figure 2d), histone H3Ser10 phosphorylation (Figure 2f), and live-cell imaging (Figure 3) showed that depletion of SIK3 increased the mitotic duration in human cell lines. Live-cell imaging analysis also indicated that mitotic exit was delayed in the absence of SIK3 (Figure 4).
A broad implication of this study is that if specific inhibitors of SIK3 can be developed, they may be able to act synergistically with conventional antimitotic drugs. Inactivation of SIK3 may allow lower doses of antimitotic drugs to be used, and thus may benefit patients by lowering the side effects of the drugs. As a test of principle, depletion of SIK3 enhanced mitotic arrest and cell death induced by classic spindle poisons, including nocodazole and Taxol (Figure 5). Downregulation of SIK3 also promoted mitotic arrest induced by newer classes of antimitotic drugs, including those targeting AURKA, AURKB, PLK1 (Figure 6), and Eg5 (Figure 7). Remarkably, siSIK3 could also restore drug sensitivity of cells resistant to the Eg5 inhibitor SB743921 (Figure 7d). Another possibility is that cancer cells with low expression of SIK3 may be more sensitive to antimitotic drugs.
It is interesting that while siSIK3 delayed mitosis mainly after the metaphase was formed, treatment with siSIK3 and antimitotic chemicals together did not necessarily delay cells after metaphase. For example, treatment of cells with siSIK3 and the Eg5 inhibitor SB743921 together arrested cells in early mitosis similar to treatment with SB743921 alone (Figure 7c), suggesting that SIK3 may have multiple roles in mitosis. Interestingly, siSIK3 did not act synergistically with HDAC inhibitors including SAHA (Figure 7e) and an HDAC6-specific inhibitor (data not shown). Although this is consistent with a model that SIK3 may act through HDACs to regulate mitosis, more refined approaches will be required in the future to test this hypothesis.
As repression of SIK3 alone resulted in a mild extension of mitosis (Supplementary Figure S2A) rather than the extensive cell death induced by inhibition of classic mitotic kinases such as PLK1 (Figure 3a), SIK3 probably may not be a very effective stand-alone target. On the other hand, it can be argued that inhibition of essential mitotic kinases such as PLK1 is too toxic for normal cells. Less critical mitotic kinases such as SIK3 may actually provide better opportunities for combined anticancer therapies.
Mitotic kinases in mammalian cells
One surprising finding of the present study is the small number of genes that overlap with the mitotic kinome using Drosophila cells.2 Of the 60 genes that affect some aspects of mitosis in Drosophila cells, only four homologs were also found to affect the mitotic index in mouse cells (Figure 1c). Bettencourt-Dias et al.2 also specifically examined the mitotic index and found six candidates (polo, for, fray, gw1, CG7156, inaC) that increase the mitotic index to outside 90% CI. Among these, only the polo kinase homolog was also found in our screen.
It is reasonable to surmise a bona fide difference of mitotic regulators between Drosophila and mammalian cells. While the mouse kinome (540 genes) is similar in size to the human kinome (518 genes),22 the Drosophila kinome is considerably smaller (228 genes). Furthermore, many of the mouse kinome siRNAs probably affected aspects of mitotic regulation but did not significantly influence mitotic duration (which was the only parameter designed to be addressed in this study). Another more trivial explanation that is inherited in most RNAi screens is the unpredictable differences in the efficiency and specificity of RNAi. Indeed, although several established mitotic kinases, such as PLK1, was identified in our screen, some notable ones including AURKA was missing. As we have subsequently found using human cell lines, knockdown of AURKA also only marginally extended mitosis (our unpublished data), suggesting that it is perhaps challenging to deplete AURKA to a level that induces mitotic delays. However, knockdown of other established mitotic kinases is expected not to increase the mitotic index, as they would either prevent entry into mitosis (e.g. CDK1) or promote mitotic slippage (e.g. AURKB).
Finally, several candidates we identified have already been linked to some aspects of mitotic control, including Erbb3,23 Lats2,24 Pim1,25 Scyl1,26 and Stk38l.27 It would be important to corroborate these and other novel candidates and establish if they are promising antimitotic targets.
In conclusion, our whole-kinome screening revealed a number of potentially interesting mitotic kinases in mammalian cells. Downregulation of SIK3, one of the conserved mitotic kinases in mammalian and Drosophila cells, delayed mitotic exit and increased the overall mitotic duration. Furthermore, depletion of SIK3 enhanced the mitotic delay and cytotoxicity induced by a number of antimitotic chemicals, highlighting the potential of SIK3 as an antimitotic drug target.
Materials and Methods
Materials
All reagents were obtained from Sigma-Aldrich (St. Louis, MO, USA), unless stated otherwise.
siRNA library screening
Mouse kinase RNAi collection was obtained from Life Technologies (Carlsbad, CA, USA). Each gene in the library was targeted by three different Stealth siRNAs. The Stealth siRNA was diluted 1 : 10 in Opti-MEM Reduced Serum Medium and transferred to 96-well plates (10 μl each well). The Stealth siRNA was mixed with 0.2 μl of Lipofectamine RNAiMAX reagent and 10 μl of Opti-MEM Reduced Serum Medium, and incubated at 25 °C for 20 min. NIH3T3/H2B-GFP cells were then added to the wells (5 × 103 cells in 100 μl of medium). The plates were incubated at 37 °C in a humidified incubator at 37 °C in 5% CO2 for 24 h. The cells were then incubated with 0.2 μg/ml of Adriamycin for 2.5 h. Images of the cells were then captured automatically using a fluorescent microscopy. For each well, bright-field and fluorescent images from nine random positions were captured. The mitotic index of each transfection was scored independently by three individuals.
Plasmid constructs
GFP-HA-SIK3 in pEGFP-C1 was obtained from the University of Dundee (Cat no. DU1338). It was cut with SacI and BamHI and ligated into pGEX-KG (GE Healthcare, Amersham, UK) to generate GST-HA-SIK3CΔ591 in pGEX-KG. GFP-HA-SIK3 in pEGFP-C1 was cut with BamHI and ligated into BamHI-cut and phosphatase-treated pUHD-P3T/PUR28 to generate FLAG-HA-SIK3 in pUHD-P3T/PUR.
Cell culture
Hep3B, HepG2, and IMR90 were obtained from the American Type Culture Collection (Manassas, VA, USA). The HeLa used in this study was a clone that expressed the tTA tetracycline repressor chimera.29 Nasopharyngeal carcinoma cell lines C666-1,30 CNE2,31 HNE1,32 and HONE132 were obtained from NPC AoE Cell Line Repository (The University of Hong Kong, Pokfulam, Hong Kong). Normal liver cells LO2 and MIHA were gifts from Irene Ng (The University of Hong Kong). Cells were propagated in RPMI1640 (for C666-1) or Dulbecco’s modified Eagle’s medium (for other cell lines) supplemented with 10% (v/v) calf serum (Life Technologies) (for HeLa) or fetal bovine serum (Life Technologies) (for other cell lines) and 50 U/ml penicillin streptomycin (Life Technologies). IMR90 cells were cultured in minimum essential medium (Eagle’s) (Life Technologies) supplemented with 2 × essential and non-essential amino acid and 15% uninactivated fetal bovine serum. Telomerase-immortalized nasopharyngeal epithelial cell lines NP361, NP460, and NP550 33 (gifts from George Tsao, The University of Hong Kong) were propagated in keratinocyte serum-free medium (Life Technologies) supplemented with 50% (v/v) Epilife. Cells were cultured in humidified incubators at 37 °C in 5% CO2. H1299,34 HCT116,34 HeLa,35 Hep3B,36 NIH3T334 that stably expressed histone H2B-GFP were used for live-cell imaging. Unless stated otherwise, cells were treated with the following reagents at the indicated final concentration: Adriamycin (0.2 μg/ml), Alisertib (Selleck Chemicals, Houston, TX, USA) (62.5 nM), Barasertib (Selleck Chemicals) (12.5 nM), MK-5108 (Selleck Chemicals) (125 nM), BI-2536 (1.25 nM), nocodazole (0.1 μg/ml), RO-3306 (Alexis) (10 μM), SB743921 (Selleck Chemicals) (10 nM), SAHA (Selleck Chemicals) (1 nM), Taxol (20 ng/ml), thymidine (2 mM), and UCN-01 (100 nM). Synchronization at mitosis was performed by first releasing cells from a double thymidine block37 for 6 h before adding nocodazole for another 6 h. Mitotic cells were then collected by mechanical shake off and released by washing three times with PBS before replating in fresh medium. To release cells from the G2 phase, HeLa were first treated with 10 μM of RO-3306 for 18 h. The cells were washed two times with PBS and replated in fresh medium. Cell-free extracts were prepared as described previously.38
Generation of SB743921-resistant cells
HeLa cells were cultured in the presence of 2.5 nM of SB743921 for 2 weeks. The cells were then subcultured with limited dilution in 5 nM of SB743921 for three weeks. Individual colonies were isolated and tested for resistance to 10 nM of SB743921-induced G2/M arrest with flow cytometry. The cells were subsequently propagated in the absence of SB743921.
Ionizing radiation
IR was delivered with a caesium137 source from an MDS Nordion (Ottawa, ON, Canada) Gammacell 1000 Elite Irradiator.
siRNA and transfection
Cells were transfected with siRNA by Lipofectamine RNAiMAX (Life Technologies). The following siRNAs were obtained from the indicated suppliers: 5′-UUCUUCGAACGUGUCACGUTT-3′ or 5′-ACGUGACACGUUCGGAGAATT-3′ (control siRNA; GenePharma, Shanghai, China); 5′-GCUGCACAAGAGGAGGAAATT-3′ (siPLK1; RiboBio, Guangzhou, China); 5′-CCACAGAAUGUGAGCAUUUTT-3′ (siSIK3 no. 1; RiboBio); 5′-CCUGAAGCAUUGGUGCGCUAUUUGU-3′ (siSIK3 no. 2; Life Technologies; Stealth siRNA); CCAAAUUGCCCUUGUGUUUTT (siSIK3 no. 3; RiboBio). Unless stated otherwise, siSIK3 no. 1 was used.
Flow cytometry
Flow cytometry analysis after propidium iodide staining was performed as described previously.39 Bivariate analysis of DNA content and histone H3Ser10 phosphorylation was performed as described previously.40
Live-cell imaging
The setup and conditions of time-lapse microscopy of living cells were as described previously.34
Antibodies and immunological methods
Antibodies against β-actin,41 CDK1,42 cyclin B1,35 and FLAG tag43 were obtained from sources as described previously. Antibodies against phospho-histone H3Ser10 and securin (sc-56207) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). GST-HA-tagged SIK3CΔ591 was expressed in bacteria and purified with GSH-agarose chromatography. Rabbit antibodies against SIK3 were raised against a bacterially expressed GST-HA-tagged SIK3CΔ591 protein.44 Immunoblotting and immunoprecipitation were performed as described.38
Abbreviations
- AMPK:
-
AMP-activated protein kinase
- APC/C:
-
anaphase-promoting complex/cyclosome
- HDAC:
-
histone deacetylase
- PLK1:
-
polo-like kinase 1
- SIK3:
-
salt-inducible kinase 3
References
Ma HT, Poon RY . How protein kinases co-ordinate mitosis in animal cells. Biochem J 2011; 435: 17–31.
Bettencourt-Dias M, Giet R, Sinka R, Mazumdar A, Lock WG, Balloux F, Zafiropoulos PJ, Yamaguchi S, Winter S, Carthew RW, Cooper M, Jones D, Frenz L, Glover DM . Genome-wide survey of protein kinases required for cell cycle progression. Nature 2004; 432: 980–987.
Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S . The protein kinase complement of the human genome. Science 2002; 298: 1912–1934.
Wang Z, Takemori H, Halder SK, Nonaka Y, Okamoto M . Cloning of a novel kinase (SIK) of the SNF1/AMPK family from high salt diet-treated rat adrenal. FEBS Lett 1999; 453: 135–139.
Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, Hawley SA, Udd L, Makela TP, Hardie DG, Alessi DR . LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J 2004; 23: 833–843.
Al-Hakim AK, Goransson O, Deak M, Toth R, Campbell DG, Morrice NA, Prescott AR, Alessi DR . 14-3-3 cooperates with LKB1 to regulate the activity and localization of QSK and SIK. J Cell Sci 2005; 118: 5661–5673.
Walkinshaw DR, Weist R, Kim GW, You L, Xiao L, Nie J, Li CS, Zhao S, Xu M, Yang XJ . The tumor suppressor kinase LKB1 activates the downstream kinases SIK2 and SIK3 to stimulate nuclear export of class IIa histone deacetylases. J Biol Chem 2013; 288: 9345–9362.
Wang B, Moya N, Niessen S, Hoover H, Mihaylova MM, Shaw RJ, Yates JRr, Fischer WH, Thomas JB, Montminy M . A hormone-dependent module regulating energy balance. Cell 2011; 145: 596–606.
Uebi T, Itoh Y, Hatano O, Kumagai A, Sanosaka M, Sasaki T, Sasagawa S, Doi J, Tatsumi K, Mitamura K, Morii E, Aozasa K, Kawamura T, Okumura M, Nakae J, Takikawa H, Fukusato T, Koura M, Nish M, Hamsten A, Silveira A, Bertorello AM, Kitagawa K, Nagaoka Y, Kawahara H, Tomonaga T, Naka T, Ikegawa S, Tsumaki N, Matsuda J, Takemori H . Involvement of SIK3 in glucose and lipid homeostasis in mice. PLoS One 2012; 7: e37803.
Sasagawa S, Takemori H, Uebi T, Ikegami D, Hiramatsu K, Ikegawa S, Yoshikawa H, Tsumaki N . SIK3 is essential for chondrocyte hypertrophy during skeletal development in mice. Development 2012; 139: 1153–1163.
Wehr MC, Holder MV, Gailite I, Saunders RE, Maile TM, Ciirdaeva E, Instrell R, Jiang M, Howell M, Rossner MJ, Tapon N . Salt-inducible kinases regulate growth through the Hippo signalling pathway in Drosophila. Nat Cell Biol 2013; 15: 61–71.
Charoenfuprasert S, Yang YY, Lee YC, Chao KC, Chu PY, Lai CR, Hsu KF, Chang KC, Chen YC, Chen LT, Chang JY, Leu SJ, Shih NY . Identification of salt-inducible kinase 3 as a novel tumor antigen associated with tumorigenesis of ovarian cancer. Oncogene 2011; 30: 3570–3584.
Manfredi MG, Ecsedy JA, Chakravarty A, Silverman L, Zhang M, Hoar KM, Stroud SG, Chen W, Shinde V, Huck JJ, Wysong DR, Janowick DA, Hyer ML, Leroy PJ, Gershman RE, Silva MD, Germanos MS, Bolen JB, Claiborne CF, Sells TB . Characterization of Alisertib (MLN8237), an investigational small-molecule inhibitor of aurora A kinase using novel in vivo pharmacodynamic assays. Clin Cancer Res 2011; 17: 7614–7624.
Marxer M, Ma HT, Man WY, Poon RYC . p53 deficiency enhances mitotic arrest and slippage induced by pharmacological inhibition of Aurora kinases. Oncogene 2013 e-pub ahead of print 19 August 2013; doi:10.1038/onc.2013.325.
Shimomura T, Hasako S, Nakatsuru Y, Mita T, Ichikawa K, Kodera T, Sakai T, Nambu T, Miyamoto M, Takahashi I, Miki S, Kawanishi N, Ohkubo M, Kotani H, Iwasawa Y . MK-5108, a highly selective Aurora-A kinase inhibitor, shows antitumor activity alone and in combination with docetaxel. Mol Cancer Ther 2010; 9: 157–166.
Yang J, Ikezoe T, Nishioka C, Tasaka T, Taniguchi A, Kuwayama Y, Komatsu N, Bandobashi K, Togitani K, Koeffler HP, Taguchi H, Yokoyama A . AZD1152, a novel and selective aurora B kinase inhibitor, induces growth arrest, apoptosis, and sensitization for tubulin depolymerizing agent or topoisomerase II inhibitor in human acute leukemia cells in vitro and in vivo. Blood 2007; 110: 2034–2040.
Steegmaier M, Hoffmann M, Baum A, Lenart P, Petronczki M, Krssak M, Gurtler U, Garin-Chesa P, Lieb S, Quant J, Grauert M, Adolf GR, Kraut N, Peters JM, Rettig WJ . BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol 2007; 17: 316–322.
Sakowicz R, Finer JT, Beraud C, Crompton A, Lewis E, Fritsch A, Lee Y, Mak J, Moody R, Turincio R, Chabala JC, Gonzales P, Roth S, Weitman S, Wood KW . Antitumor activity of a kinesin inhibitor. Cancer Res 2004; 64: 3276–3280.
Cadot B, Brunetti M, Coppari S, Fedeli S, de Rinaldis E, Dello Russo C, Gallinari P, De Francesco R, Steinkuhler C, Filocamo G . Loss of histone deacetylase 4 causes segregation defects during mitosis of p53-deficient human tumor cells. Cancer Res 2009; 69: 6074–6082.
Eot-Houllier G, Fulcrand G, Magnaghi-Jaulin L, Jaulin C . Histone deacetylase inhibitors and genomic instability. Cancer Lett 2009; 274: 169–176.
Richon VM, Webb Y, Merger R, Sheppard T, Jursic B, Ngo L, Civoli F, Breslow R, Rifkind RA, Marks PA . Second generation hybrid polar compounds are potent inducers of transformed cell differentiation. Proc Natl Acad Sci USA 1996; 93: 5705–5708.
Caenepeel S, Charydczak G, Sudarsanam S, Hunter T, Manning G . The mouse kinome: discovery and comparative genomics of all mouse protein kinases. Proc Natl Acad Sci USA 2004; 101: 11707–11712.
Wang Q, Greene MI . EGFR enhances Survivin expression through the phosphoinositide 3 (PI-3) kinase signaling pathway. Exp Mol Pathol 2005; 79: 100–107.
Tao W, Zhang S, Turenchalk GS, Stewart RA, St John MA, Chen W, Xu T . Human homologue of the Drosophila melanogaster lats tumour suppressor modulates CDC2 activity. Nat Genet 1999; 21: 177–181.
Bachmann M, Hennemann H, Xing PX, Hoffmann I, Moroy T . The oncogenic serine/threonine kinase Pim-1 phosphorylates and inhibits the activity of Cdc25C-associated kinase 1 (C-TAK1): a novel role for Pim-1 at the G2/M cell cycle checkpoint. J Biol Chem 2004; 279: 48319–48328.
Kato M, Yano K, Morotomi-Yano K, Saito H, Miki Y . Identification and characterization of the human protein kinase-like gene NTKL: mitosis-specific centrosomal localization of an alternatively spliced isoform. Genomics 2002; 79: 760–767.
Hergovich A, Stegert MR, Schmitz D, Hemmings BA . NDR kinases regulate essential cell processes from yeast to humans. Nat Rev Mol Cell Biol 2006; 7: 253–264.
Ma HT, Tsang YH, Marxer M, Poon RY . Cyclin A2-cyclin-dependent kinase 2 cooperates with the PLK1-SCFbeta-TrCP1-EMI1-anaphase-promoting complex/cyclosome axis to promote genome reduplication in the absence of mitosis. Mol Cell Biol 2009; 29: 6500–6514.
Yam CH, Siu WY, Lau A, Poon RY . Degradation of cyclin A does not require its phosphorylation by CDC2 and cyclin-dependent kinase 2. J Biol Chem 2000; 275: 3158–3167.
Cheung ST, Huang DP, Hui AB, Lo KW, Ko CW, Tsang YS, Wong N, Whitney BM, Lee JC . Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein–Barr virus. Int J Cancer 1999; 83: 121–126.
Sizhong Z, Xiukung G, Yi Z . Cytogenetic studies on an epithelial cell line derived from poorly differentiated nasopharyngeal carcinoma. Int J Cancer 1983; 31: 587–590.
Glaser R, Zhang HY, Yao KT, Zhu HC, Wang FX, Li GY, Wen DS, Li YP . Two epithelial tumor cell lines (HNE-1 and HONE-1) latently infected with Epstein–Barr virus that were derived from nasopharyngeal carcinomas. Proc Natl Acad Sci USA 1989; 86: 9524–9528.
Li HM, Man C, Jin Y, Deng W, Yip YL, Feng HC, Cheung YC, Lo KW, Meltzer PS, Wu ZG, Kwong YL, Yuen AP, Tsao SW . Molecular and cytogenetic changes involved in the immortalization of nasopharyngeal epithelial cells by telomerase. Int J Cancer 2006; 119: 1567–1576.
On KF, Chen Y, Ma HT, Chow JP, Poon RY . Determinants of mitotic catastrophe on abrogation of the G2 DNA damage checkpoint by UCN-01. Mol Cancer Ther 2011; 10: 784–794.
Chan YW, Ma HT, Wong W, Ho CC, On KF, Poon RY . CDK1 inhibitors antagonize the immediate apoptosis triggered by spindle disruption but promote apoptosis following the subsequent rereplication and abnormal mitosis. Cell Cycle 2008; 7: 1449–1461.
Ma HT, Chan YY, Chen X, On KF, Poon RY . Depletion of p31comet protein promotes sensitivity to antimitotic drugs. J Biol Chem 2012; 287: 21561–21569.
Ma HT, Poon RY . Synchronization of HeLa cells. Methods Mol Biol 2011; 761: 151–161.
Poon RY, Toyoshima H, Hunter T . Redistribution of the CDK inhibitor p27 between different cyclin.CDK complexes in the mouse fibroblast cell cycle and in cells arrested with lovastatin or ultraviolet irradiation. Mol Biol Cell 1995; 6: 1197–1213.
Siu WY, Arooz T, Poon RY . Differential responses of proliferating versus quiescent cells to adriamycin. Exp Cell Res 1999; 250: 131–141.
Chow JP, Siu WY, Ho HT, Ma KH, Ho CC, Poon RY . Differential contribution of inhibitory phosphorylation of CDC2 and CDK2 for unperturbed cell cycle control and DNA integrity checkpoints. J Biol Chem 2003; 278: 40815–40828.
Chan YW, On KF, Chan WM, Wong W, Siu HO, Hau PM, Poon RY . The kinetics of p53 activation versus cyclin E accumulation underlies the relationship between the spindle-assembly checkpoint and the postmitotic checkpoint. J Biol Chem 2008; 283: 15716–15723.
Siu WY, Lau A, Arooz T, Chow JP, Ho HT, Poon RY . Topoisomerase poisons differentially activate DNA damage checkpoints through ataxia-telangiectasia mutated-dependent and -independent mechanisms. Mol Cancer Ther 2004; 3: 621–632.
Fung TK, Siu WY, Yam CH, Lau A, Poon RY . Cyclin F is degraded during G2–M by mechanisms fundamentally different from other cyclins. J Biol Chem 2002; 277: 35140–35149.
Harlow E, Lane DP . Antibodies: A Laboratory Manual Cold Spring Harbor Laboratory Press: New York, USA, 1988.
Acknowledgements
We thank Nelson Lee for help in generating the SIK3 antibodies and Charlie Forecar for technical assistance. This work was supported in part by the Research Grants Council 662213, HKU7/CRG/09 and AOE-MG/M-08/06 to RYCP.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by M Agostini
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Chen, H., Huang, S., Han, X. et al. Salt-inducible kinase 3 is a novel mitotic regulator and a target for enhancing antimitotic therapeutic-mediated cell death. Cell Death Dis 5, e1177 (2014). https://doi.org/10.1038/cddis.2014.154
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2014.154
Keywords
This article is cited by
-
Biomarker discovery in attention deficit hyperactivity disorder: RNA sequencing of whole blood in discordant twin and case-controlled cohorts
BMC Medical Genomics (2020)
-
The potent roles of salt-inducible kinases (SIKs) in metabolic homeostasis and tumorigenesis
Signal Transduction and Targeted Therapy (2020)
-
Targeting LKB1 in cancer – exposing and exploiting vulnerabilities
British Journal of Cancer (2015)