Abstract
Menin, encoded by the multiple endocrine neoplasia type 1 ( MEN1 ) gene, is a tumor suppressor that leads to multiple endocrine tumors upon loss of its function. Menin functions as a transcriptional activator by tethering MLL complex to mediate histone H3 K4 methylation. It also functions as a repressor. However, the molecular mechanism of how menin contributes to the opposite outcome in gene expression is largely unknown. Here, we investigated the role of menin in the epigenetic regulation of transcription mediated by histone covalent modification. We show that the global methylation level of histone H3 K9, as well as H3 K4, was decreased in Men1 −/− MEF cells. Consistently, menin was able to interact with the suppressor of variegation 3–9 homolog family protein, SUV39H1, to mediate H3 K9 methylation. This interaction decreased when patient-derived MEN1 mutation was introduced into the SUV39H1-interaction domain. We show that menin mediated different chromatin changes depending on target genes. Chromatin immunoprecipitation studies showed that menin directly associated with the GBX2 promoter and menin-dependent recruitment of SUV39H1 was essential for chromatin remodeling and transcriptional regulation. These results provide a molecular basis of how menin functions as a transcriptional repressor and suggest that menin-dependent integration of H3 K9 methylation might play an important role in preventing tumors.
Similar content being viewed by others
Main
Menin is a tumor suppressor protein encoded by the multiple endocrine neoplasia type 1 (MEN1) gene,1 and its loss of function due to diverse missense and nonsense mutations along the entire gene is observed in either inheritable or sporadic neoplasm of multiple endocrine organs including the parathyroid gland, pituitary gland, and pancreatic islets.2 A mouse model shows that menin is an essential protein whose absence results in embryonic lethality, whereby heterozygous mice develop endocrine tumors, consistent with its role as a classic tumor suppressor for the endocrine lineage.3, 4
Menin does not share any specific motifs or detectable sequence similarities with any other known proteins, which makes the functional study of menin complicated. Therefore, identification of proteins that interact with menin has significantly contributed to deciphering its biological roles. Menin has been reported to engage in a complex network of interactions with a broad spectrum of proteins, including transcription factors: JunD, Pem, NF-(κ)B, Smad3, β-catenin, and the mixed lineage leukemia (MLL) complex; proteins involved in regulation of DNA repair: RPA2 and FANCD2; kinases: ASK and nm23H1; and cytoskeletal proteins: non-muscle myosin heavy chain IIA, glial fibrillary acidic protein, and Vimentin.5 The diversity of interacting partners suggests roles for menin in multiple biological pathways, including gene transcription, cell cycle control, apoptosis, genome stability, and hematopoiesis.6, 7 However, the precise molecular mechanism of menin as a tumor suppressor needs to be further investigated.
Histone proteins that form nucleosomes within chromatin engage in diverse cellular processes via extensive post-translational modifications such as acetylation and methylation. Methylation of histone subunits on lysine residues is one of the relatively well-established epigenetic mechanisms.8, 9 For example, methylation of histone H3 K4 is associated with transcriptional activation,10, 11 while histone H3 K9 methylation is, in general, related to transcriptional silencing.12, 13, 14 Histone H3 lysine methylation is catalyzed by different histone methyltransferases (HMTs) with evolutionarily conserved SET domains; multiprotein assemblies with homology to the Drosophila trithorax complex and the related yeast Set1 complex methylate histone H3 on K4, while HMTs, such as SUV39H1 and G9a, have H3 K9 methylation activity.15, 16, 17, 18, 19, 20, 21 It is of note that not only the positions of the lysine residues, but also the numbers of methyl groups on the same residue affect molecular consequences of gene expression.
Several lines of investigation indicate that the function of menin is tightly connected to chromatin function. Menin is known to serve as both an activator and repressor of transcription via integration of different chromatin modification complexes. For example, menin mediates repression of genes targeted by the transcription factor JunD through mSin3A-histone deacetylase (HDAC) complex22, 23 and the growth factor pleiotrophin gene through polycomb group complex-mediated histone H3 K27 methylation.24 In contrast, the expression of genes encoding cyclin-dependent kinase (CDK) inhibitors, p27Kip1 and p18INK4c, is promoted by menin through the interaction with MLL–HMT complex that mediates histone H3 K4 methylation.25, 26, 27, 28 As the epigenetic regulation of transcription has been recognized as a major mechanism of gene regulation in eukaryotic cells, it continuously gathers attention on how menin contributes to the epigenetic regulation of gene expression and the relevance to its role in tumorigenesis.
In this study, we sought to investigate the role of menin in epigenetic regulation of transcription through the integration of a variety of histone codes. Our data show that menin has an ability to interact with different classes of HMTs via distinct domains. We demonstrate that menin interacts with SUV39H1 to repress expression of target genes such as GBX2. Menin/SUV39H1 complex contained HDAC1, implying transcriptional repression via either histone methylation or histone deacetylation, or both. Menin may function as a tumor suppressor by regulating histone methylation states at the promoters of specific target genes that govern proliferation and tumorigenesis.
Results
Menin interacts with SUV39H1
The Men1+/− islets were reported to reduce the global level of H3 K4 trimethylation.26 To investigate a more broad range of epigenetic impact of menin in tumor suppressor function, we tested if menin affects other histone modifications as well as H3 K4 methylation. We confirmed that H3 K4 trimethylation level was reduced in Men1−/−MEF cells compared with that in Men1+/+MEF cells (Figure 1a). In addition to H3 K4 methylation, interestingly, we found a profoundly decreased level of H3 K9 trimethylation in Men1−/−MEF cells, whereas H3 K4 dimethylation, H3 K9 dimethylation, and H3 K14 acetylation levels showed no remarkable changes (Figure 1a, left panel). To test whether H3 K4 and K9 trimethylation are specifically affected by menin, Men1−/−cells were infected with a control vector or a vector expressing menin. The expression of menin in the infected cells was confirmed by western blot analysis and immunohistochemistry (Figure 1a and Supplementary Figure S1). Complementation of cells with menin restored the H3 K4 and -K9 trimethylation significantly but it hardly affected the levels of H3 K4 and -K9 dimethylation or H3 K14 acetylation (Figure 1a, right panel), indicating that menin specifically affects methylation of histone H3 K9, as well as H3 K4. To further examine the effect of menin on histone modification, MEF cells were treated with pan-HDAC inhibitor, TricostatinA (TSA). H3 K9 is one of histone residues competitively modified by acetylation for activation or methylation for repression. The acetylation of several different K residues was increased by TSA treatment in both Men1+/+and Men1−/− MEF cells, but H3 K9 acetylation (and the H3 acetylation) was remarkably more greatly increased in Men1−/−MEF cells (Figure 1b). This dramatic increase was not observed in the cases of H3 K4, K14, and H4 K12 acetylation. Our data suggest the possibility that menin leads to a significant amount of H3 K9 methylation that might efficiently compete out the acetylation on the same residue.
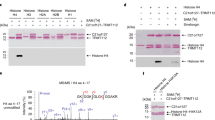
Menin specifically associates with SUV39H1. (a) Whole-cell lysates of Men1+/+and Men1−/−MEFs were used for western blot analysis with antibodies against menin, H3K4me2, H3K4me3, H3K9me2, H3K9me3, H3K14ac, H3, and GAPDH (left panel). H3 and GAPDH served as loading controls. Men1−/− MEF cells were infected with control or menin-expressing retroviruses. Cell lysates from the infected cells were subjected to western blot analysis (right panel). (b) Menin’s effect on histone covalent modification. MEF cells were treated with TSA (100 nM) for 24 h and whole-cell extract was prepared for western blot analysis. H3, Tubulin, and GAPDH are loading controls. (c) 293T cells were transfected with Flag-SUV39H1, Flag-HDAC1, and Myc-menin expression vectors as indicated. Whole-cell extracts were immunoprecipitated with anti-Flag antibody, and the immunoprecipitates were analyzed by western blotting using anti-Flag and anti-Myc antibodies. (d) Co-immunoprecipitation assay was performed to detect interaction between endogenous proteins in 293T cells. Anti-SUV39H1 antibody was used for immunoprecipitation. The asterisks * denote cross-reactive IgG heavy chain bands. As SUV39H1 co-migrates with and is masked by IgG heavy chain on SDS-PAGE, the supernatant fraction remained after IP is shown in parallel to show that SUV39H1 was efficiently precipitated by its antibody. (e) In vitro HMT activity associated with menin. HMT reaction was performed with menin immunoprecipitates and [3H]-SAM. Each reaction contains bacterially expressed GST-H3N (residues 1–57) wild-type (wt) or mutants (K4R, K4R/K27R and K4R/K9R/K27R) as indicated. Proteins were resolved on a SDS-polyacrylamide gel. The gel was stained with Coomassie (bottom) and exposed to film for fluorography (top)
As menin affects the level of H3 K9 methylation, we searched for a potential HMT that mediates menin-dependent H3 K9 methylation. In this effort, we examined whether menin associates with SUV39H1, a specific HMT that targets H3 K9. 293T cells were transfected with expression vectors for Myc-menin and Flag-SUV39H1. Whole cell lysates were subjected to immunoprecipitation (IP) with anti-Flag antibody and the IP pellets were analyzed for the presence of Myc-menin by western blot analysis. A menin-interacting protein, HDAC1, was used as a positive control. Myc-menin was detected when Flag-SUV39H1 was co-expressed (Figure 1c), indicating that menin associates with SUV39H1. Furthermore, IP with antibody against SUV39H1 showed that menin interacted with SUV39H1 at endogenous protein level in 293T cells (Figure 1d). In this condition, as immunoprecipitated SUV39H1was masked by the IgG heavy chain, it was indirectly confirmed that IP was successful by showing that SUV39H1 was completely depleted in the supernatant. We next analyzed menin-mediated H3 K9 methylation activity. Endogenous menin was immunoprecipitated by anti-menin antibody and was subjected to the in vitro HMT assay. This assay included GST-fused N terminus of histone H3 (wild type or mutants with K4, K9, and K27 residues respectively substituted by arginine) as substrates. GST-H3N was methylated only when K9 was present (Figure 1e). Taken together, these data suggest that menin has an ability to interact with SUV39H1 and potentially influences H3 K9 methylation in vivo.
SUV39H1 interaction domain is mapped on the distinct region of menin
To determine the domain within menin that mediates interaction with SUV39H1, we performed IP from extracts of 293T cells transfected with GFP-SUV39H1 and various versions of truncated menin with the Myc tag. The N-terminal truncation mutants maintained their interaction with SUV39H1 until they lost almost more than a half of the protein from the N-terminus (Figure 2a and Supplementary Figure S2), whereas further deletion beyond amino acid 360 (ΔN445) failed to interact with SUV39H1, indicating that the region encompassing amino acids 360–445 (SvID; SUV39H1 interaction domain) is involved in SUV39H1 interaction. In comparison, MLL interaction domain was recently resolved in detail from the structural analysis of menin to the central cavity that provides the binding surface for MLL.28 Our domain mapping data and the structure of menin/MLL complex indicate that distinct domains are involved in SUV39H1 and MLL interactions (Figure 2b). The SvID seems to contain a bundle of α-helixes, located toward the C-terminus from the concave. As SUV39H1 has an independent ability to interact with HDAC1,29 menin, SUV39H1, and HDAC1 were co-precipitated within complex by IP (Supplementary Figure S3).
Mapping of HMT-binding domains in menin. (a) 293T cells were transfected with Myc-menin- and GFP-SUV39H1-expressing vectors. Whole-cell extracts were immunoprecipitated with anti-Myc antibody, and the immunoprecipitates were analyzed by western blotting with antibodies against GFP, or Myc epitope. The asterisks * denote nonspecific cross-reactive bands. (b) Schematic diagram showing association of SUV39H1 with the indicated versions of menin. MLL-binding region is denoted. (c) Each35[S]-labeled menin mutant derived from MEN1 patients was mixed with Flag-SUV39H1-expressing cell lysates and subjected to IP with anti-Flag antibody. Proteins were resolved by SDS-PAGE, and their interaction was detected by autoradiography. (A, alanine; P, proline; D, aspartic acid; N, asparagine; L, leucine; W, tryptophan; R, arginine)
The importance of the SvID was further tested using menin point mutants, which were reported in MEN1 patients. The four mutants, A411P, L413P, D418N, and W436R, carrying substitution within the SvID, were labeled in vitro with 35[S]-methionine along with wild-type menin. IP was performed by incubating labeled proteins with partially purified Flag-tagged SUV39H1. As shown in Figure 2c, D418N and W436R were affected in the interaction with SUV39H1. Our data indicate that menin interacts with SUV39H1 and this ability might be involved in its tumor suppressor function. Interestingly, we and others have previously reported that parafibromin, one of the human PAF1 complex subunits, recruits SUV39H1 and downregulates cyclinD1 gene.30, 31 As parafibromin and menin have common cellular (endocrine tumor suppressor) and molecular (SUV39H1 interaction) functions, we compared two regions mapped for SUV39H1 interaction, by comparative protein structure modeling with threading method.32 Despite the difference in amino acid sequences, the SUV39H1 binding domains of parafibromin and menin appear to have similar folds with the helix-loop-helix structure (Supplementary Figure S4).
Menin and SUV39H1 have common targets for gene regulation
To investigate the contribution of menin to gene regulation via H3 K9 methylation or SUV39H1, we performed DNA microarray analysis using mRNAs isolated from Men1−/−and Men1+/+ MEF cells. A substantial number of genes were elevated or reduced in the Men1−/−MEFs as expected. Among genes whose arbitrary difference in expression was 1.2-fold or greater, 2814 transcripts were identified as upregulated in the Men1−/−cells (Supplementary Figure S6). In an effort to find potential SUV39H1 target genes, we also performed DNA microarray analysis with a set of Men1+/+ MEF cells transfected with either siRNA control (scrambled) or siRNA targeting SUV39H1 (Supplementary Figure S5). It showed that expression of 2599 genes was increased more than 1.2-fold by depletion of SUV39H1 (Figure 3a and Supplementary Figure S6). Approximately 649 genes were identified overlapping with the genes whose expression was elevated by lack of menin, implying that these might represent a subset of genes potentially co-regulated by menin and SUV39H1. Gene ontology analysis revealed that many of the affected genes were involved in cell metabolism, signal transduction, cell cycling, immunity and defense, and development (Supplementary Figure S6). Among them, of particular interest is the GBX2 homeobox gene, which is known to be consistently overexpressed in cancer cells.33, 34 The homeobox genes including GBX2 and HLXB9 are related to normal development and function of the pancreas.35 Furthermore, activation of HLXB9 by loss of menin has been proposed to contribute to islet beta cell tumorigenesis.36 We validated microarray data by performing qRT-PCR, showing that the GBX2 mRNA level was significantly increased in menin-null cells (Figure 3b). To further test, a menin-depleted condition was established in Men1+/+ cells by Men1-specific siRNA (Supplementary Figure S5). As anticipated, Men1-knockdown increased GBX2 mRNA level (Figure 3c). In contrast, reconstitution of Men1−/−cells with wild-type menin reduced the expression of GBX2 (Figure 3d). The direct role of SUV39H1 on GBX2 expression was analyzed using cells treated with siRNA to inhibit the expression of SUV39H1. SUV39H1 siRNA did not affect the levels of menin, and vice versa (Supplementary Figure S5). Notably, depletion of SUV39H1 significantly increased the level of GBX2 mRNA in Men1+/+MEF cells, but not in Men1−/− MEF cells (Figure 3e). Furthermore, although individual depletion of menin and SUV39H1 increased the expression of GBX2, simultaneous depletion of both proteins did not have an additive effect on GBX2 level (Figure 3f), indicating that menin and SUV39H1 function in the same pathway in a cross-dependent manner. Taken together, our data show that SUV39H1 and menin function together at the protein level and lead to the downregulation of GBX2.
Menin and SUV39H1 contribute to GBX2 repression. (a) The cDNA microarray analysis of MEF cells reveals common targets of menin and SUV39H1. Commonly upregulated genes by depletion of menin (Men−/−/Men+/+) and SUV39H1 (SUV39H1 siRNA/control) were indicated. Expression of GBX2 was measured using qRT-PCR in Men1+/+ or Men1−/− MEFs (b) as well as in MEFs that had been treated with Men1 siRNA (c). (d) The level of GBX2 mRNA was analyzed in Men1−/− MEFs infected with retroviral vectors expressing empty (control) or menin. (e) Knockdown of SUV39H1 increased the mRNA level of GBX2 in Men1+/+MEF cells, but not in Men1−/− MEF cells. Men1+/+ and Men1−/− MEF cells were treated for 1 day with siRNA specifically targeting SUV39H1 and total RNA was isolated to detect the steady state level of GBX2 by qRT-PCR. (f) Double knockdown of both menin and SUV39H1 did not have an additive effect on GBX2 mRNA level. The PCR values were normalized for GAPDH and presented as relative values by considering GBX2/GAPDH of control as 1. Error bars represent S.D., n=3. Significance of differences was evaluated (*P-value<0.05, **P-value<0.005)
Epigenetic regulation mediated by menin depends on the context
As menin is known to tether HDAC activity to the target genes to repress their expression, we sought to examine the relative contribution of menin-dependent HDAC and HMT activities to the gene regulation. Insulin-like growth factor-binding protein 2 (IGFBP2) gene was previously known to be repressed by menin through its promoter.37, 38 However, the molecular mechanism was not identified clearly. Our genome-wide analysis revealed IGFBP2 is upregulated by menin depletion but not changed by SUV39H1 depletion. We thus focused on IGFBP2 gene as a menin target regulated independently of SUV39H1. We confirmed that the expression of the IGFBP2 promoter-linked reporter gene was repressed by menin (Figure 4a). The mRNA level of endogenous IGFBP2 was negatively affected by menin, but it was unaltered by the treatment of SUV39H1 siRNA (Figure 4b and c), indicating that IGFBP2 is regulated by menin in a SUV39H1-independent manner.
Epigenetic regulation of IGFBP2 is mediated by menin via HDAC. (a) Menin represses IGFBP2 promoter activity. The 293T cells were transfected with the IGFBP2 promoter-luciferase reporter and epitope-tagged menin or empty vector control. After incubation for 24 h, luciferase activity was measured. Each transfection was performed in duplicate and repeated three times. The means±S.D. of duplicate determinations from three separate experiments are shown. The expression level of transfected menin is shown at the bottom. (b) Increased level of genomic IGFBP2 expression in Men1−/− MEFs was confirmed using qRT-PCR. (c) Men1+/+ MEFs cells were treated with siRNA targeting SUV39H1. The expression level of IGFBP2 was analyzed by qRT-PCR. Error bars represent S.D., n=3. Significance of differences was evaluated (*P-value<0.05). (d and e) Men1+/+ and Men1−/− MEF cells were treated with TSA for 24 h with increasing dose as indicated. Total RNA was prepared to perform qRT-PCR. The expression level of IGFBP2 and GBX2 were normalized by GAPDH. TSA-dependency was shown as the fold difference between samples by considering expression levels obtained from each Men1+/+ and Men1−/− MEF cells without TSA treatment as 1, respectively. Error bars represent S.D., n=3 (*P-value<0.05)
To examine the relative contribution of menin-mediated HDAC activity to the regulation of IGFBP2 or GBX2, we next treated Men1+/+ and Men1−/−MEF cells with TSA. The contribution of HDAC activity can be monitored by TSA-dependency. For this, the mRNA level was normalized by GAPDH and the value obtained without TSA treatment in each MEF cells was arbitrarily set to 1. TSA-treated samples were compared by relative fold change. As shown in Figure 4d, the expression of IGFBP2 was greatly increased by the treatment of TSA in a dose-dependent manner in the presence of menin, whereas the fold increase by TSA was greatly reduced when menin was absent, indicating that menin contributed to IGFBP2 regulation through the HDAC activity. The expression of GBX2 was also increased by TSA, implying the involvement of HDAC activity. However, overall induction fold of GBX2 and the dependency on TSA was small and relatively similar in Men1−/− and Men1+/+ MEF cells, compared with that of IGFBP2 (Figure 4e). Our data indicate that the repression of IGFBP2 is mediated largely by menin-dependent coupling of HDAC activity, while SUV39H1 is more critical for the menin-dependent regulation of GBX2. Hence, these results indicate that menin might exert different epigenetic influence depending on the genetic context.
Menin recruits SUV39H1 to promote chromatin remodeling at the GBX2 locus
To investigate the regulatory mechanism of GBX2 by menin and whether SUV39H1 has any role in chromatin structure, we performed ChIP with Men1+/+ and Men1−/−MEF cells using anti-menin antibody. We used two distinct pairs of primers for ChIP analysis, each targeting the GBX2 promoter and 3′-end region (Figure 5a). Our ChIP data show that menin bound to the GBX2 promoter in Men1+/+ cells, but not in Men1−/− cells (Figure 5b). These results provide evidence that menin is involved in GBX2 expression by direct binding to the GBX2 promoter. We then tested the possibility that SUV39H1 was recruited to the GBX2 gene through the interaction with menin by comparing its occupancy in the presence and absence of menin. ChIP with anti-SUV39H1 showed the association of SUV39H1 with GBX2 promoter region in Men1+/+MEF cells, which, however, decreased in Men1−/− MEF cells (Figure 5c). To elucidate a potential mechanism by which menin mediates GBX2 regulation, we analyzed the effect of menin on several histone modifications. Interestingly, menin significantly affected H3 K9 methylation. Both H3 K9 tri- and dimethylation were reduced in Men1−/− cells, whereas the levels of H3 K4 tri- and dimethylation were increased (Figures 5d and e). Although we did not observe remarkable changes in global H3 K9 dimethylation with bulk histones (see Figure 1a), ChIP analysis within GBX2 locus revealed local changes in H3 methylation, indicating that menin contributes to formation of overall repressive chromatin environment, which is in part via SUV39H1. This result is expected as SUV39H1 is preferentially a trimethylase of H3 K9 and also a dimethylase to a lesser extent.39 Menin is known to interact with MLL; however, MLL was hardly detected in GBX2 promoter by ChIP in both cell types (Supplementary Figure S7), in contrast to its occupancy at HoxA9 region,40 indicating that menin might not target MLL for the regulation of GBX2. Consistent with the decrease of H3 K9 methylation, H3 K9 acetylation was increased, reflecting the elevated transcription of GBX2 (Figure 5f). Furthermore, when the promoter and 3′-end regions of GBX2 were compared, changes in histone modification were prominent in the promoter that menin occupied (Figures 5d–f). Our data suggest that SUV39H1 is recruited to the GBX2 promoter via interaction with menin to induce H3 K9 trimethylation, thus providing the repressive chromatin environment for downregulation of GBX2.
Menin recruits SUV39H1 to promote trimethylation of H3K9 around the GBX2 locus. (a) GBX2 locus showing regions subjected to ChIP analysis. (b and c) Recruitment of menin and SUV39H1 along the GBX2 locus was analyzed by ChIP assay. Chromatin solution was prepared from Men1+/+ and Men1−/− cells and subjected to ChIP using control rabbit IgG and the antibodies against endogenous menin (anti-menin Ab) (b) or SUV39H1(anti-SUV39H1 Ab) (c). Immunoprecipitated DNA was analyzed in duplicates by quantitative real-time PCR with primers shown above (a) and the relative enrichment of proteins was shown. (d–f) ChIP assay with Men1+/+ and Men1−/− MEFs using antibodies against indicated H3 modifications was performed to measure the relative levels of H3 K4 or H3 K9 di-, trimethylation, and acetylation along the GBX2. The data represent the percentage of ChIP (IP/input) and error bars indicate the S.D., n=3. Significance of differences was evaluated (*P-value<0.05)
Discussion
Human menin is a 615-amino acid protein with a distinctive concave structure that is comprised of an internal core domain and two flanking α-helical bundles. Menin is peculiar in that it functions as a tumor suppressor in endocrine tissues and an oncogenic cofactor in blood cells by playing within a complex protein–protein interaction network, which must be context-dependent or cell type-specific. One of the key interacting partners of menin is MLL, a H3 K4 HMT. Menin and MLL mediate transcriptional activation of CDK inhibitor genes in pancreas and the HOX genes during development.26, 27, 40 In addition, menin functions as a transcriptional repressor as it interacts with the mSin3/HDAC1 complex to mediate JunD-dependent transcriptional repression.22 Here, we show that menin has an independent ability to integrate SUV39H1 for gene repression. Approximately 649 genes (∼23% of downregulated genes by menin) including GBX2 are potential candidates that are likely to be influenced either directly or indirectly by menin and SUV39H1. Therefore, we propose that menin uses multiple ways for epigenetic regulation of target genes in the different context.
Gbx2 is a transcription factor of homeobox family proteins that function in cell identity and differentiation and reported to be consistently elevated in human prostate cancer tissues.33, 34 One of the known downstream targets of Gbx2, Interleukin 6 (IL-6), stimulates autocrine/paracrine signaling loop through the activation of STAT3 pathway to confer tumorigenic properties.41 Although the role of Gbx2 in MEN1 development has not yet been described, cytokines such as IL-6 are also implicated in endocrine tumors as their expression levels correlate with the tumorigenic and metastatic potential.42, 43 In this study, we found that GBX2 is a common target of menin and SUV39H1 from the analysis of non-biased genome-wide gene expression of MEF cells by showing that it is de-repressed upon deletion of menin or SUV39H1. Our data show that the interaction of menin and SUV39H1 is important for induction of H3 K9 methylation at the GBX2 promoter. Although menin was known to mediate H3 K4 methylation through the MLL complex for expression of CDK inhibitors or HOX genes, menin might target particularly H3 K9 methylation of GBX2 and subsequently suppress inflammation and tumor growth in the endocrine lineage. In this model, given the multiple interaction partners with chromatin modifying activity, menin might be able to function as a transcriptional activator through MLL complex or a repressor through HDAC/SUV39H1 in different target genes, which provide an efficient epigenetic regulatory mechanism for cell cycle and cell tumorigenesis (Figure 6). It is not clear whether HDAC activity tethered by menin is a prerequisite for the H3 K9 methylation. Two types of histone modification could occur in an independent or a synergistic manner depending on gene contexts.
A model for a dual role of menin in gene regulation during tumorigenic pathways. Menin may function as a transcriptional activator or repressor in a context-dependent manner, by selective mediation of chromatin remodeling, which provides an efficient mechanism for the regulation of gene expression and cell tumorigenesis
Interestingly, our results are reminiscent of the parafibromin (the human homolog of yeast Cdc73) subunit of the human PAF1 complex, consisting of Paf1, parafibromin, Leo1, Rtf1, Ctr9, and Ski8, which have both positive and negative roles in transcription.44, 45 The PAF1 complex is required for H3 methylation (K4 and K79) and H2B ubiquitination, thus coordinating transcriptional events and chromatin remodeling which is important for HOX gene expression.46, 47 In particular, such parafibromin also mediates SUV39H1 binding, via a protein domain whose predicted secondary structure is similar to that of menin, to repress cyclinD1 and c-myc gene through H3 K9 methylation.30, 48 Of special note, parafibromin is a tumor suppressor associated with endocrine tumors.49 Many sporadic and germline mutations of HRPT2 that encodes parafibromin result in loss of function and tumor development.
Then what is the underlying mechanism that allows menin to function as a transcriptional activator or repressor? And how is it related to menin’s dual role as a tumor suppressor and an oncogenic driver? As menin’s transcriptional property is likely to be determined at least partly by interacting partners, there might be a regulatory mechanism that influences the nature of the protein interaction. In case of parafibromin, SHP2-dependent status of the phosphorylation on tyrosine residue of parafibromin is important to shift its transcriptional activity between activation and repression by influencing interaction with β-catenin or SUV39H1.31 Likewise, menin is found phosphorylated in vivo,50 providing a possibility that menin’s opposing transcriptional activities could be switched by upstream signaling pathways that govern interacting partners depending on the phosphorylation status, although it is not yet clear whether phosphorylation has any regulatory role in menin’s function. Further work to identify a full list of interacting partners and their cognate critical targets and a work to disintegrate the dynamic nature of the interacting network will give more insights to understanding menin’s role in gene expression during anti- and pro-oncogenic processes.
Materials and methods
Plasmids
pcDNA3-Menin-Myc/His,22 pCDNA3 Flag-SUV39H1,51 pBJ5 Flag-HDAC1,22 pIGFBP2-Luc,37 and pEGFP-C1-SUV39H130 were previously described. DNA constructs expressing menin truncation mutants were generated by PCR and inserted into pcDNA3-Myc/His vector (Invitrogen, Carlsbad, CA, USA). Patient-derived missense mutants of menin were constructed by PCR-mediated site-directed mutagenesis and confirmed by sequencing.
Cell culture, transfection, and siRNAs
293T cells were cultured in Dulbecco’s Modified Eagles’ Medium (DMEM) supplemented with 10% fetal bovine serum and 50 units per ml of penicillin/streptomycin (HyClone, Waltham, MA, USA). Men1+/+ MEF and Men1−/−MEF cells were cultured in DMEM with 10% fetal bovine serum, 50 units per ml of penicillin/streptomycin, 2 mM L-glutamine (HyClone), and 0.1 mM non-essential amino acids (Welgene, Daegu, Korea). 293T cells were transfected with transfection reagents according to the manufacturer’s protocol (Bio-Rad, Berkeley, CA, USA). For RNA interference assay, cells were transfected with a control siRNA (AccuTarget negative control siRNA, Bioneer, Daejeon, Korea) or siRNAs against SUV39H1 (Samchully, Seoul, Korea or Santa Cruz Biotechnology, Santa Cruz, CA, USA) and Men1 (Samchully, Korea) using Lipofectamine 2000 (Invitrogen).
Western blot analysis, immunoprecipitation
For western blotting, cells were lysed in lysis buffer (20 mM TrisCl (pH 7.4), 150 mM NaCl, 0.5% NP-40, and inhibitors of protease and phosphatase). The extracted proteins were resolved by SDS-PAGE and transferred onto nitrocellulose membranes. For immunoprecipitation, 293T cells were harvested 24 h after transfection and lysed with lysis buffer. Thus obtained lysates were immunoprecipitated with appropriate antibodies along with protein-A/G beads (Amersham Pharmacia Biotech, GE Healthcare, Buckinghamshire, UK). Immunoprecipitates were resolved by SDS-PAGE, transferred onto nitrocellulose membranes, and subjected to immunoblotting with appropriate antibodies.
In vitro HMT assays
Immunoprecipitation of menin was performed as described above. The immunoprecipitates were washed four times with IP Buffer (20 mM TrisCl (pH 7.4), 150 mM NaCl, 0.5% NP-40) and once more with HMT Buffer (50 mM Tris (pH 8.5), 20 mM KCl, 10 mM MgCl2, 10 mM β-mercaptoethanol, and 250 mM sucrose). Each reaction contained 1 μl of S-adenosyl-L-(methyl-3H) methionine (Perkin Elmer, Waltham, MA, USA) and GST-fused N terminus of histone H3 (GST-H3N) that was expressed and purified from Escherichia coli strain BL21. Reactions were incubated for 2 h at 30 °C. Sample buffer was added and samples were boiled for 5 min to stop the reaction. Proteins were fractionated on SDS-PAGE, stained with Coomassie blue and analyzed by fluorography.
In vitro 35[S]-labeling
The deletion mutants of menin were synthesized and labeled with 35[S]-methionine using STP3 (T7) in vitro transcription/translation kit (Promega, Madison, WI, USA). In vitro-labeled mutants were incubated with cell lysates obtained from 293T cells expressing Flag-tagged SUV39H1 for 4 h in the presence of anti-Flag antibody and protein-G beads. Immunoprecipitates were subjected to SDS-PAGE and analyzed by autoradiography.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Men1+/+ and Men1−/− MEF cells were transfected with control, menin, or SUV39H1 siRNAs for 24 h. Total RNA was isolated using Trizol (Invitrogen), and cDNA synthesis was performed with the Reverse Transcription System (Fermentas, Waltham, MA, USA). Quantitative real-time PCR was performed with specific primers for GBX2 and GAPDH using the KAPA SYBRFAST qPCR KIT (Kapa Biosystems, Woburn, MA, USA) and CFX96 real-time PCR detector (Bio-Rad). Relative levels of mRNA were normalized to the values of GAPDH mRNA for each reaction. The following primers were used: GBX2 (#1483; 5′-CAGATGACAATTTGCCTGGTCAG-3′ and #1484; 5′-CAGGGAGAGGTACTTTTTGCAGTG-3′), IGFBP2 (#1178; 5′-CCTGGAACATCTCTACTCCCTGCAC-3′ and #1179; 5′-CCAGTCTCCTGCTGCTCGTTG-3′), SUV39H1 (#1424; 5′-TTGGCTGTGAGTGCCAGGAC-3′ and #1425; 5′-CTTTCTGGACTACACGGTTTGGG-3′), MEN1(#1343; 5′-TCACAAGGGAATTGCCTCAGC-3′ and #1344; 5′-GGCCACTTCAAAGAATTCCTTGTAG-3′), and GAPDH (#790; 5′-AACATCAAATGGGGTGAGGCC-3′ and #791; 5′-GTTGTCATGGATGACCTTGGC-3′).
DNA microarray
Men1+/+ and Men1−/− MEF cells were seeded in a 150-mm dish. The cells were cultured for 24 h before total RNA was prepared using RNeasy Plus Mini kit (Qiagen, Valencia, CA, USA). After processing with DNase digestion and clean-up procedures, quantified RNA samples were amplified and purified using the Ambion Illumina RNA amplification kit (Ambion, Austin, TX, USA) to yield biotinylated cRNA according to the manufacturer’s instructions. Labeled cRNAs were hybridized to each mouse-8 expression bead array (Illumina Inc., San Diego, CA, USA) and the array signal was detected using Amersham fluorolink streptavidin-Cy3 (GE Healthcare Bio-Sciences, Little Chalfont, UK). Arrays were scanned with an Illumina Bead Array Reader Confocal Scanner and array data export processing and analysis was performed using Illumina BeadStudio v3.1.3 (Gene expression Module v3.3.8). Differentially expressed genes were detected on the basis of fold-change threshold (1.2-fold) and P-value (<0.05). Microarray analysis with RNA samples prepared from siRNA-treated Men1+/+ MEFs followed the same procedure.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as previously reported according to the manufacturer’s protocol (Upstate Biotechnology, Gaffney, SC, USA).30 Sonicated lysates were used for ChIP with immunoprecipitated DNA and input DNA were analyzed by quantitative real-time PCR using specific primers; GBX2 promoter (#1491; 5′-TGTCGTGTGAGCAAGGGCTG-3′ and #1492; 5′-CATGAGAGGCGGCTTGGAAC-3′), GBX23′ end region (#1495; 5′-CTTGCATGTCCTCTTTTTGCATGT-3′ and #1496; 5′-TGCTTTTAGGGTCGGAGTTTCTTC-3′), Hoxa9 (#2109; 5′-AATGCGATTTGGCTGCTTTTTTATGGC-3′ and #2110; 5′-TCAAATCTGGCCTTGCCTCTG-3′).
Antibodies
Antibodies included: anti-menin (Bethyl Laboratories, Montgomery, TX, USA), anti-SUV39H1 (Abcam (Cambridge, UK) or Upstate), anti-GAPDH, anti-α-Tubulin (LabFrontier, Anyang, Korea), anti-H3 K4 2me, anti-H3 K9 2me, anti-H3 K9 3me, anti-acetyl H3 K14 (Millipore, Billerica, MA, USA), anti-acetyl H3 K9 (Cell Signaling, Boston, MA, USA), anti-acetyl H3 (Millipore), anti-acetyl H4 (Upstate), anti-acetyl H3 K4 (Active Motif, Carlsbad, CA, USA), anti-acetyl H4 K12 (Abcam), anti-H3 and anti-H3 K4 3me (Abcam), anti-Myc, and anti-Flag (M2) (Sigma Aldrich, St Louis, MO, USA).
Abbreviations
- ChIP:
-
chromatin immunoprecipitation
- GBX2:
-
gastrulation brain homeobox 2
- HDAC:
-
histone deacetylase
- HMT:
-
histone methyltransferase
- HOX:
-
homeobox
- IGFBP2:
-
insulin-like growth factor binding protein-2
- IL-6:
-
interleukin 6
- IP:
-
immunoprecipitation
- MEF:
-
mouse embryo fibroblast
- MLL:
-
mixed lineage leukemia
- qRT-PCR:
-
quantitative reverse transcription-polymerase chain reaction
- siRNA:
-
small interfering RNA
- TSA:
-
trichostatin A
References
Chandrasekharappa SC, Guru SC, Manickam P, Olufemi SE, Collins FS, EmmertBuck MR et al. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science 1997; 276: 404–407.
Chandrasekharappa SC, Teh BT . Functional studies of the MEN1 gene. J Intern Med 2003; 253: 606–615.
Bertolino P, Radovanovic I, Casse H, Aguzzi A, Wang ZQ, Zhang CX . Genetic ablation of the tumor suppresor menin causes lethality at mid-gestation with defects in multiple organs. Mech Dev 2003; 120: 549–560.
Crabtree JS, Scacheri PC, Ward JM, Garrett-Beal L, Emmert-Buck MR, Edgemon KA et al. A mouse model of multiple endocrine neoplasia, type 1, develops multiple endocrine tumors. Proc Natl Acad Sci USA 2001; 98: 1118–1123.
Yang Y, Hua X . In search of tumor suppressing functions of menin. Mol Cell Endocrinol 2007; 265: 34–41.
Lemos MC, Thakker RV . Multiple endocrine neoplaslia type 1 (MEN 1): analysis of 1336 mutations reported in the first decade following identification of the gene. Hum Mutat 2008; 29: 22–32.
Balogh K, Patocs A, Hunyady L, Racz K . Menin dynamics and functional insight: take your partners. Mol Cell Endocrinol 2010; 326: 80–84.
Jenuwein T, Allis CD . Translating the histone code. Science 2001; 293: 1074–1080.
Turner BM . Cellular memory and the histone code. Cell 2002; 111: 285–291.
Noma K, Grewal SIS . Histone H3 lysine 4 methylation is mediated by Set1 and promotes maintenance of active chromatin states in fission yeast. Proc Natl Acad Sci USA 2002; 99: 16438–16445.
Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci USA 2002; 99: 8695–8700.
Nakayam J, Rice JC, Strahl BD, Allis CD, Grewal SIS . Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 2001; 292: 110–113.
Peters A, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 2001; 107: 323–337.
Noma K, Allis CD, Grewal SIS . Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 2001; 293: 1150–1155.
Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SYR et al. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev 2001; 15: 3286–3295.
Goo YH, Sohn YC, Kim DH, Kim SW, Kang MJ, Jung DJ et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol 2003; 23: 140–149.
Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M et al. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci USA 2001; 98: 12902–12907.
Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML . A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci USA 2002; 99: 90–94.
Roguev A, Schaft D, Shevchenko A, Pijnappel W, Wilm M, Aasland R et al. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. Embo J 2001; 20: 7137–7148.
Choi J, Jang H, Kim H, Kim ST, Cho EJ, Youn HD . Histone demethylase LSD1 is required to induce skeletal muscle differenitation by regulating myogenic factors. Bioch Biophy Res Comm 2010; 401: 327–332.
Rea S, Eisenhaber F, O’Carroll N, Strahl BD, Sun ZW, Schmid M et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000; 406: 593–599.
Kim H, Lee JE, Cho EJ, Liu JO, Youn HD . Menin, a tumor suppressor, represses JunD-mediated transcriptional activity by association with an mSin3A-histone deacetylase complex. Cancer Res 2003; 63: 6135–6139.
Kim H, Lee JE, Kim BY, Cho EJ, Kim ST, Youn HD . Menin represses JunD transcriptional activity in protein kinase C-mediated Nur77 expression. Exp Mol Med 2005; 37: 466–475.
Gao SB, Feng ZJ, Xu B, Wu Y, Yin P, Yang Y et al. Suppression of lung adenocarcinoma through menin and polycomb gene-mediated repression of growth factor pleiotrophin. Oncogene 2009; 28: 4095–4104.
Milne TA, Hughes CM, Lloyd R, Yang ZH, Rozenblatt-Rosen O, Dou YL et al. Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors. Proc Natl Acad Sci USA 2005; 102: 749–754.
Karnik SK, Hughes CM, Gu XY, Rozenblatt-Rosen O, McLean GW, Xiong Y et al. Menin regulates pancreatic islet growth by promoting histone methylation and expression of genes encoding p27(Kip1) and p18(INK4c). Proc Natl Acad Sci USA 2005; 102: 14659–14664.
Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC et al. Menin associates with a trithorax family histone methyltransferase complex and with the Hoxc8 locus. Mol Cell 2004; 13: 587–597.
Huang Jing, Gurung Buddha, Wan Bingbing, Matkar Smita, Veniaminova Natalia A, Wan Ke et al. The same pocket in menin binds both MLL and JUND but has opposite effects on transcription. Nature 2012; 482: 542–546.
Vaute O, Nicolas E, Vandel L, Trouche D . Functional and physical interaction betwee the histone mehtyl transferase Suv39H1 and histone deacetylases. Nucleic Acids Res 2002; 30: 475–481.
Yang YJ, Han JW, Youn HD, Cho EJ . The tumor suppressor, parafibromin, mediates histone H3 K9 methylation for cyclin D1 repression. Nucleic Acids Res 2010; 38: 382–390.
Takahashi A, Tsutsumi R, Kikuchi I, Obuse C, Saito Y, Seidl A et al. SHP2 tyrosine phosphatase converts parafibromin/Cdc73 from a tumor suppressor to an oncogenic driver. Mol Cell 2011; 43: 45–56.
Kelley LA, Sternberg MJE . Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 2009; 4: 363–371.
Gao AC, Lou W, Isaacs JT . Enhanced GBX2 expression stimulates growth of human prostate cancer cells via transcriptional up-regulation of the interleukin 6 gene. Clin Cancer Res 2000; 6: 493–497.
Gao AC, Lou W, Isaacs JT . Down-regulation of homeobox gene GBX2 inhibits human prostate cancer clonogenic ability and tumorigenicity. Cancer Res 1998; 58: 1391–1394.
Mizusawa N, Hasegawa T, Ohigashi I, Tanaka-Kosugi C, Harada N, Itakura M et al. Differentiation phenotypes of pancreatic islet beta- and alpha-cells are closely related with homeotic genes and a group of differentially expressed genes. Gene 2004; 331: 53–63.
Scacheri PC, Davis S, Odom DT, Crawford GE, Perkins S, Halawi MJ et al. Genome-wide analysis of menin binding provides insights into MEN1 tumorigenesis. PLoS Genet 2006; 2: 406–419.
La P, Schnepp RW, Petersen CD, Silva AC, Hua XX . Tumor suppressor menin regulates expression of insulin-like growth factor binding protein 2. Endocrinology 2004; 145: 3443–3450.
La P, Desmond A, Hou Z, Silva AC, Schepp RW, Hua X . Tumor suppressor menin: the essential role of nuclear localization signal domains in coordinating gene expression. Oncogene 2006; 25: 3537–3546.
Robin P, Fritsch L, Philipot O, Svinarchuk F, Ait-Si-Ali S . Post-translational modifications of histones H3 and H4 associated with the histone methyltransferases Suv39h1 and G9a. Genome Biol 2007; 8: R270.
Chen YX, Yan JZ, Keeshan K, Tubbs AT, Wang HR, Silva A et al. The tumor suppressor menin regulates hematopoiesis and myeloid transformation by influencing Hox gene expression. Proc Natl Acad Sci USA 2006; 103: 1018–1023.
Ni Z, Lou W, Leman ES, Gao AC . Inhibition of constitutively activated stat3 signaling pathway suppresses growth of prostate cancer cells. Cancer Res 2000; 60: 1225–1228.
Okitsu K, Kanda T, Imazeki F, Yonemitsu Y, Ray RB, Chang C et al. Involvement of interleukin-6 and androgen receptor signaling in pancreatic cancer. Genes Cancer 2010; 1: 859–867.
Lesina M, Kurkowski MU, Ludes K, Rose-John S, Treiber M, Klöppel G et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell 2011; 19: 456–469.
Chaudhary K, Deb S, Moniaux N, Ponnusamy MP, Batra SK . Human RNA polymerase II-associated factor complex: dysregulation in cancer. Oncogene 2007; 26: 7499–7507.
Jaehning JA . The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim Biophys Acta Gene Regul Mech 2010; 1799: 379–388.
Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P et al. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell 2005; 20: 601–611.
Wood A, Schneider J, Dover J, Johnston M, Shilatifard A . The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem 2003; 278: 34739–34742.
Lin L, Zhang JH, Panicker LM, Simonds WF . The parafibromin tumor suppressor protein inhibits cell proliferation by repression of the c-myc proto-oncogene. Proc Natl Acad Sci USA 2008; 105: 17420–17425.
Carpten JD, Robbins CM, Villablanca A, Forsberg L, Presciuttini S, Bailey-Wilson J et al. HRPT2, encoding parafibromin, is mutated in hyperparathyroidism-jaw tumor syndrome. Nature Genet 2002; 32: 676–680.
Francis J, Lin WC, Rozenblatt-Rosen O, Meyerson M . The menin tumor suppressor protein is phosphorylated in response to DNA damage. PLoS One 2011; 6: 16119.
Jang H, Choi DE, Kim H, Cho EJ, Youn HD . Cabin1 represses MEF2 transcriptional activity by association with a methyltransferase, SUV39H1. J Biol Chem 2007; 282: 11172–11179.
Acknowledgements
We thank Dr. Sunita K Agarwal (NIH, Bethesda, MD, USA) and Dr. Chang-Xian Zhang (Centre National de la Recherché Scientifique-Unite Mixte de Recherché, University of Lyon, Lyon, France), Ming Lei (University of Michigan Medical School), Xianxin Hua (University of Pennsylvania) for providing plasmids and Dr. Hwangseo Park (Sejong University) for analysis of protein structure. This work was supported by Mid-career Researcher Program (ROA-2008-0060084 and 2009-0080410) and Basic Science Research Program (2010–0028646) through the National Research Foundation of Korea grant funded by the Ministry of Education, Science and Technology to E-JC.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by D Aberdam
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Yang, YJ., Song, TY., Park, J. et al. Menin mediates epigenetic regulation via histone H3 lysine 9 methylation. Cell Death Dis 4, e583 (2013). https://doi.org/10.1038/cddis.2013.98
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.98
Keywords
This article is cited by
-
Abnormal expression of menin predicts the pathogenesis and poor prognosis of adult gliomas
Cancer Gene Therapy (2020)
-
Deletion of Menin in craniofacial osteogenic cells in mice elicits development of mandibular ossifying fibroma
Oncogene (2018)
-
SUV39H1 mediated SIRT1 trans-repression contributes to cardiac ischemia–reperfusion injury
Basic Research in Cardiology (2017)
-
The Treatment Landscape and New Opportunities of Molecular Targeted Therapies in Gastroenteropancreatic Neuroendocrine Tumors
Targeted Oncology (2017)
-
Towards a new classification of gastroenteropancreatic neuroendocrine neoplasms
Nature Reviews Clinical Oncology (2016)