Abstract
In human cells, the RIPK1–RIPK3–MLKL–PGAM5–Drp1 axis drives tumor necrosis factor (TNF)-induced necroptosis through mitochondrial fission, but whether this pathway is conserved among mammals is not known. To answer this question, we analyzed the presence and functionality of the reported necroptotic axis in mice. As in humans, knockdown of receptor-interacting kinase-3 (RIPK3) or mixed lineage kinase domain like (MLKL) blocks TNF-induced necroptosis in L929 fibrosarcoma cells. However, repression of either of these proteins did not protect the cells from death, but instead induced a switch from TNF-induced necroptosis to receptor-interacting kinase-1 (RIPK1) kinase-dependent apoptosis. In addition, although mitochondrial fission also occurs during TNF-induced necroptosis in L929 cells, we found that knockdown of phosphoglycerate mutase 5 (PGAM5) and dynamin 1 like protein (Drp1) did not markedly protect the cells from TNF-induced necroptosis. Depletion of Pink1, a reported interactor of both PGAM5 and Drp1, did not affect TNF-induced necroptosis. These results indicate that in these murine cells mitochondrial fission and Pink1 dependent processes, including Pink-Parkin dependent mitophagy, apparently do not promote necroptosis. Our data demonstrate that the core components of the necrosome (RIPK1, RIPK3 and MLKL) are crucial to induce TNF-dependent necroptosis both in human and in mouse cells, but the associated mechanisms may differ between the two species or cell types.
Similar content being viewed by others
Main
Necroptosis is a form of regulated necrosis that strictly depends on the kinase activity of receptor-interacting kinase-1 (RIPK1) and receptor-interacting kinase-3 (RIPK3).1 This cell death modality operates independently of executioner caspases, but is negatively regulated by caspase-8 and therefore is sensitized under conditions of caspase-8 inhibition or deficiency.2 Necroptosis was first observed in L929 murine fibrosarcoma cells as a regulated form of cell death.2 But only with the recent availability of necrostatins3 and RIPK3 knockout mice4 was its importance noted in various pathologies, including ischemia/reperfusion,5, 6 inflammatory bowel disease,7, 8 viral infection,9 Salmonella infection10 and sepsis.11
The involvement of functional necroptosis in vivo greatly relies on the use of RIPK1 kinase inhibitors such as necrostatins3, 5 and the discovery of RIPK3 as a decisive pro-necroptotic kinase.9, 12, 13 Members of the tumor necrosis factor (TNF) family are potent inducers of necroptosis. TNF-induced necroptosis involves the formation of a necrosome complex consisting of the core components RIPK1, RIPK3 and mixed lineage kinase domain like (MLKL) that are negatively regulated by factors such as Fas associated death domain protein (FADD), caspase-8 and cellular FLICE inhibitory protein.1, 14 Despite the importance of necroptosis, its molecular components and the mechanisms of its regulation and execution remain elusive. Until recently, the only known downstream substrates of RIPK1 and RIPK3 have been RIPKs serving as their own substrates.9 But last year two novel RIPK3 substrates were reported: mixed lineage kinase domain like (MLKL)15, 16 and phosphoglycerate mutase 5 (PGAM5).17
MLKL was independently identified by two different groups, who showed that it is constitutively bound by a wild type but not by the kinase-dead RIPK3.15, 16 During TNF-induced necroptosis, RIPK3 phosphorylates human MLKL at positions T357 and S358, and these phosphorylations were shown to be essential for TNF-induced necroptosis.15 Although Zhao et al.16 identified MLKL by screening a short hairpin library against kinases/phosphatases in HT-29 cells, Sun et al.15 identified MLKL by screening a large library of compounds against TNF-necroptosis in HT-29 cells and showed that necrosulfonamide (NSA) inhibits TNF-driven necroptosis. Biotinylated NSA was subsequently used to pull down and identify MLKL.15 Importantly, the Cys86 residue in human MLKL – which is targeted by NSA – is absent in murine MLKL.15 Consequently, NSA does not delay necroptosis in murine cells.15
The second reported RIPK3 substrate is PGAM5.17 In humans, the PGAM5 gene encodes two isoforms, PGAM5-L and PGAM5-S, produced by alternative splicing.18 PGAM5 constitutively translocates to the mitochondria and has phosphatase activity, but other PGAM members involved in glucose metabolism do not have these properties.19 The phosphorylation of PGAM5 during TNF-driven necroptosis has been shown to require RIPK3.17 In turn, phosphorylated PGAM5 activates the mitochondrial fission protein, dynamin related kinase-1 (Drp1), by dephosphorylating S637, which then allows Drp1-driven mitochondrial fission.17 It has thus been proposed that RIPK3 activates the MLKL–PGAM5–Drp1 axis during necroptosis. The observed mitochondrial fission would thereby serve as a potential execution mechanism during TNF-driven necroptosis.17
In this study, we chose to further examine the contribution of the components of this novel17 axis in a prototype murine model of necroptosis. We also included Pink1, as this protein is reported to interact with PGAM520 as well as Drp1,21 influence cell death22 and affect mitochondrial fission.23 Pink1 also regulates the removal of damaged mitochondria in a process called mitophagy.24 This cellular function requires the E3 ubiquitin ligase Parkin, a downstream regulator of Pink1.25 Therefore, we studied a possible contribution of Parkin in TNF-induced necroptosis as well.
Overall, our data show that knockdown of RIPK1, RIPK3 or MLKL strongly attenuates TNF-induced necroptosis in murine cells. In contrast, repression of PGAM5, Pink1 or Parkin has no effect on necroptosis induction, and Drp1 knockdown only mildly delays TNF-induced necroptosis. These data indicate that neither mitochondrial fission nor mitophagy contribute to the execution of TNF-induced necroptosis in our murine cellular system. Of interest, absence of RIPK3 or MLKL not only blocks necroptosis but also shifts the response to RIPK1 kinase-dependent apoptosis.
Results
Knockdown of RIPK3 or MLKL blocks TNF-induced necroptosis and reveals a switch to apoptosis that is dependent on RIPK1 kinase activity
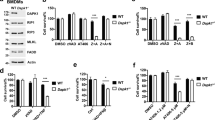
The RIPK1–RIPK3–MLKL–PGAM5–Drp1 axis operates during TNF/zVAD-fmk-induced necroptosis in the presence of SMAC mimetics (SM) in human HT-29 cells and HeLa cells transfected with RIPK3.15 It is unknown whether this pathway is conserved among mammalian cells and to which extent it influences induced apoptosis. The murine L929 fibrosarcoma cell line has been the prototype cellular model system for identifying pathways involved in necroptosis.2, 9, 12, 13, 26, 27 It allows TNF-induced necroptosis without the need for adding IAP inhibitors such as SM, or caspase inhibitors such as zVAD-fmk. Therefore, we studied the contribution of the recently identified axis to TNF-induced necroptosis in murine L929 cells. In addition, we also studied its possible contribution to anti-Fas-induced apoptosis. To identify potential cell death modality switches upon interference with either cell death pathway, we simultaneously monitored cell death and caspase activity as a function of time. Knockdown of none of the members of the RIPK1–RIPK3–MLKL–PGAM5–Drp1 axis affected Fas-induced apoptosis, which was visualized by cleavage of a synthetic caspase substrate (Figures 1a and b, right axis) and disruption of the plasma membrane (Figures 1a and b, left axis). As we showed in a previous study,14 knockdown of RIPK1 (Figure 1c) did not block TNF-induced cell death but shifted cell death from TNF-induced necroptosis to apoptosis (Figure 1a). Knockdown of MLKL (Figure 1d) significantly delayed TNF-induced necroptosis in L929 cells 6 h after TNF stimulation (Figure 1a), as previously reported for L929 cells.15, 16 Interestingly, we now show that this delayed cell death is associated with caspase activity (Figures 1a and b), indicating a shift to apoptosis. Apoptosis was confirmed by detecting chromatin condensation and plasma membrane blebbing (Figure 2a). Knockdown of RIPK3 (Figure 1c) completely blocked TNF-induced necroptosis until 6 h after stimulation (Figure 1a). However, cell death observed later was also associated with an increased caspase activity (Figure 1b), indicating a switch to apoptosis as well. Importantly, this TNF-induced apoptosis observed upon knockdown of RIPK3 or MLKL was reduced in the presence of the RIPK1 kinase inhibitor necrostatin-1 (Figure 2b), demonstrating its dependence on the kinase activity of RIPK1.
Transient knockdown of components along the RIPK1–RIPK3–MLKL–PGAM5–Drp1 axis in L929sahFas cells. Knockdown of RIPK1, RIPK3 or MLKL causes a switch from TNF-induced necroptosis to apoptosis. Seventy-two hours after knockdown of L929sahFas cells with the indicated siRNAs, cells were stimulated in triplicate with TNF (10 000 U/ml) or in duplicate with agonistic anti-Fas Ab (250 ng/ml). (a) The 6-h time point is shown±S.E.M (n=3). Cell death and caspase activity were determined as described in Materials and Methods. *P<0.05 and **P<0.01. (b) The 12-h time point is shown±S.E.M (n=3). Cell death and caspase activity were determined as described in Materials and Methods. *P<0.05. (c) Seventy-two hours after knockdown of murine RIPK1, RIPK3, Drp1 or Parkin, the knockdown efficiency of these targets was validated by western blotting, and (d) 72 h after knockdown of murine PGAM5, MLKL and Pink1, the knockdown efficiency of these targets was validated by RT-PCR because detection by antibody was unreliable
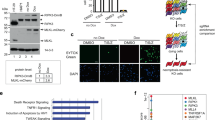
Knockdown of RIPK3 or MLKL results in delayed cell death that switches from TNF-induced necroptosis to a delayed, Nec-1 sensitive apoptosis. (a) Seventy-two hours after knockdown of L929sahFas cells with the indicated siRNAs, cells were stimulated with TNF (10 000 U/ml) or agonistic anti-Fas Ab (250 ng/ml) in the presence of 1 μg/ml HOECHST and 3 μg/ml propidium iodide (PI). Imaging was performed on a BDPathwayTM 855 (n=2). Chromatin condensation is indicated with arrows and plasma membrane blebbing with arrowheads. Scale bar indicates 20 μm. (b) Seventy-two hours after knockdown of L929sahFas cells with the indicated siRNAs, cells were pretreated with 10 μM necrostatin-1 (N1) for 1 h, followed by TNF stimulation (10 000 U/ml) in triplicate. Cell death and caspase activity were determined as described in Materials and Methods. The 12-h time point is shown±S.E.M (n=3)
Evaluation of the possible contribution of the PGAM5–Drp1 axis in TNF-induced necroptosis in L929 cells
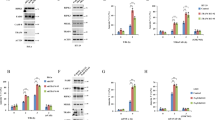
Remarkably, and in contrast to TNF-induced necroptosis in HeLa cells transfected with RIPK3,17 knockdown of PGAM5 (Figure 1d) did not influence necroptosis in L929 cells (Figures 1a and b and Supplementary Figure S1). Knockdown of the reported PGAM5 target Drp117 was very effective (Figure 1c), but it resulted only in a mild but significant delay of TNF-necroptosis (Figure 1a and Supplementary Figure S1). Note that knockdown of Drp1 markedly slowed down cell proliferation (Figures 3a and b). This was not observed for any other protein examined in this study. We therefore addressed whether the delay in cell proliferation and/or the delay in cell death caused by Drp1 knockdown could be attributed to a decrease in mitochondrial metabolism, which is known to protect from necroptosis.13, 28, 29 However, knockdown of Drp1 did not reduce mitochondrial respiration (Figure 3c), whereas pharmacological inhibition of Drp1 using mitochondrial division inhibitor (mdivi)30 did (Figure 4a). The Drp1 inhibitor mdivi markedly delayed TNF-driven necroptosis (Figure 4b), in line with the results obtained in human cells.16 The protective effects of mdivi on TNF-induced necroptosis contrast with the mild protection observed after Drp1 knockdown (Figures 1a and b and Supplementary Figure S1). Note that this delay only occurred when mdivi was used at concentrations that also block mitochondrial respiration (Figures 4a and b).
Knockdown of Drp1 delays cell proliferation but does not reduce mitochondrial respiration. (a) Forty-eight hours after transient knockdown of L929 cells with the indicated siRNAs, cells were seeded at 10 000 cells in a 96-well plate (eight replicates), and after 24 h live cells (Hoechst+/PI-) were counted using automated imaging. Data are expressed as percentage of control±S.E.M (n=3). *P<0.05. (b) Forty-eight hours after reverse transfection of L929 cells with the indicated siRNAs, cells were seeded at 10 000 cells in a 96-well plate, and 24 h later the cells were incubated in fresh medium supplemented with SytoxGreen (1.6 μM) and Triton-X100 (0.1% w/v). Subsequently, maximal fluorescence of Sytoxgreen was determined by fluorometry. Data are expressed as percentage of control±S.E.M (n=3). *P<0.05. (c) Basal, maximal and minimal oxygen consumption rates (OCR) in L929 cells were analyzed as described in Materials and Methods. Briefly, 10 000 cells were seeded in a 96-well plate (eight replicates) for each knockdown. After 24 h, cellular oxygen consumption rates were determined. The data show the kinetics of four consecutive measurements of baseline respiration. Subsequently, the protonophore CCCP was added (12 μM) to each well to dissipate the mitochondrial membrane potential and thereby promote maximal electron transport and corresponding maximal oxygen consumption. Injection of the complex I inhibitor rotenone (Rot, 2.5 μM) and complex III inhibitor Antimycin-A (AM, 5 μM) in each well was followed by four consecutive measurements (at 5 min intervals) to determine the minimal mitochondrial respiration. Finally, the amount of viable cells (HOECHST+/PI- cells) in the microchamber of each well was determined by automated imaging. The data represent the oxygen consumption rate (pmol O2 per minute) per 1000 cells±S.E.M (n=3)
The Drp1 inhibitor mdivi inhibits TNF-induced necroptosis and mitochondrial respiration. (a) L929sahFas cells were pretreated with mdivi (0–250 μM) for 1 h and respiration was analyzed as described in Materials and Methods. After four consecutive measurements of basal respiration, the protonophore CCCP was injected (12 μM) and maximal respiration was determined. Rotenone (2.5 μM) and Antimycin-A (5 μM) were then injected to determine the mitochondrial baseline respiration. A representative experiment is shown (n=2). (b) After pretreatment with mdivi (0–250 μM), L929sahFas cells were stimulated with TNF for 6 h, and cell death was determined by flow cytometry; it is expressed as percentage of SytoxGreen positive cells control±S.E.M (n=2). *P<0.05
These data suggest that mdivi delays TNF-driven necroptosis because it inhibits mitochondrial respiration rather than mitochondrial fission. In addition, the unaffected mitochondrial respiration upon Drp1 knockdown suggests that mdivi inhibits mitochondrial respiration due to off-target effects independently from targeting Drp1. This illustrates that the effect of mdivi in this context of necroptosis cannot be confined to its inhibitory activity on Drp1. In conclusion, our data demonstrate that knockdown of PGAM5 or Drp1 hardly affects TNF-induced necroptosis in L929 cells. Indeed, kinetics of TNF-necroptosis remain intact upon knockdown of PGAM5 and are mildly delayed upon knockdown of Drp1 (Supplementary Figure S1). It remains possible that the observed delay of cell proliferation upon Drp1 knockdown contributes to this mild protection from TNF-necroptosis.
Evaluation of the possible contribution of the PGAM5–PINK1–Parkin axis in TNF-induced necroptosis
We also examined the possible contribution of Pink1, a genetic interactor of PGAM520 and Drp121 and an upstream regulator of Parkin.25 Transient knockdown of Pink1 markedly delayed TNF-necrosis without a shift toward apoptosis (Figure 1a), whereas stable knockdown of Pink1 did not affect TNF-necroptosis (Supplementary Figure S2). Given these different outcomes, we examined a possible pro-necroptotic role of Pink1 using two additional approaches. In the first approach, we found that inducible overexpression of a wild-type Pink1 that is resistant to siRNAs targeting Pink1 could not revert the protective phenotype of siRNAs targeting murine Pink1 (Supplementary Figure S3 and Supplementary Data D1). These data suggest that off-target effects of the siRNAs targeting Pink1 are responsible for their protective effects against TNF-necroptosis. In a second complementary approach, we observed no significant differences in TNF/Tak1i/zVAD-fmk-induced cell death between Pink1 KO MEFs and littermate control MEFs (Supplementary Figure S4), indicating that Pink1 has no role in necroptosis.
Finally, we found that knockdown of Parkin (Figure 1c), a downstream effector of Pink1 in the induction of mitophagy, did not affect TNF-necroptosis (Figures 1a and b and Supplementary Figure S1). Because both Pink1 and Parkin did not seem to contribute to TNF-induced necroptosis, we monitored the occurrence of mitophagy, which is preceded by depolarization of mitochondrial membranes. For this purpose, we performed live cell imaging of L929 cells expressing mitochondrially targeted GFP. Within 1–3 h after TNF stimulation, the tubular mitochondrial network of mito-GFP disintegrated in a punctate pattern (Supplementary Figure S5). Importantly, these punctate mito-GFP structures retained their mitochondrial membrane potential, assessed by staining with the mitochondrial potential dye TMRM (Supplementary Figure S5). Consequently, depolarization of mitochondrial membranes, a prerequisite for mitophagy, did not occur during TNF-necroptosis. Altogether, these data argue against the involvement of Pink1–Parkin dependent processes, including mitophagy, in regulating the execution of TNF-necroptosis.
Discussion
We used a murine model of necroptosis to study the contribution of components of the recently identified RIPK1–RIPK3–MLKL–PGAM5–Drp1 axis17 promoting mitochondrial fission during TNF-induced necroptosis. We also examined whether Pink1, a genetic interactor of PGAM520 and Drp1,21 known to affect mitochondrial fragmentation,23 influences necrotic cell death.
Following TNF stimulation, different types of complexes are formed.31 Complex I mediates NF-κB activation and MAPK signaling, resulting in inflammation and cell survival. The cytosolic complex II initiates FADD-caspase-8-mediated apoptosis, which under cIAP-depleting conditions can be either independent of the RIPK1 kinase activity (so-called complex IIa) or dependent on it (so-called complex IIb). Typically, RIPK1/3 are inactivated by caspase-8-mediated cleavage, resulting in suppression of necroptosis. When caspase-8 is inhibited or absent, complex II initiates necroptosis, which is dependent on RIPK1–RIPK3–MLKL.15 Uniquely in L929 fibrosarcoma cells, TNF directly induces necroptosis without the need to block caspase-8, which is functionally present.2 We previously have shown that depletion of RIPK1 shifts TNF-induced necroptosis to an accelerated form of apoptosis that is not inhibited by Nec-1 as its target (RIPK1) is depleted.14 These results demonstrate that RIPK1 negatively controls FADD-caspase-8 mediated apoptosis. As TRADD and RIPK1 can compete for TNFR-1 binding in complex I, the absence of RIPK1 probably facilitates the interaction of pro-apoptotic proteins, including TRADD, with the TNFR1 and the formation of complex IIa.32, 33, 34, 35 Here, we show that depletion of RIPK3 or MLKL also shifts TNF-induced necroptosis to apoptosis, albeit with a delay. These results may indicate that RIPK3 and MLKL, like RIPK1, negatively control FADD-caspase-8-mediated apoptosis. Alternatively, RIPK3 and MLKL may just ensure efficient pro-necroptotic signaling, and in their absence apoptosis becomes the default outcome of triggering a cell death pathway. We also observed that the delayed apoptosis upon depletion of RIPK3 or MLKL can be inhibited by Nec-1. RIPK1-kinase-dependent apoptosis was also observed in the presence of IAP inhibitors and TNF,36 etoposide,37, 38 TRAIL39 or in the presence of Tak1 inhibitor and TNF.14, 40 These data confirm that RIPK1 kinase activity is involved in both apoptosis and necroptosis. This switch from necroptosis to RIPK1-mediated apoptosis after depletion of RIPK3 and MLKL, at least in L929 fibrosarcoma cells, should be taken into consideration when developing strategies for interfering with necroptosis induction. Note that the difference in induction rate of apoptosis between RIPK1 and RIPK3/MLKL knockdown may be due to the contribution of RIPK1 within complex I to NF-κB activation, resulting in defective induction of cell death inhibitory proteins such as Bcl-2 members and IAPs, leading to cell death ‘without brakes’.
Downstream of the RIPK1–RIPK3–MLKL axis, PGAM5 and Drp1 might function as executioners of necroptosis by activating mitochondrial fission.16 However, knockdown of PGAM5 did not alter TNF-necroptosis in the murine L929 necroptosis system, in contrast to human cells.17 Our results are in agreement with the recent report of Murphy et al.41, who showed that stable knockdown of PGAM5 did not affect TNF-driven necroptosis in both L929 and MEF cells. One possibility is that PGAM5 is a stable protein, and its knockdown at the RNA level does not guarantee the absence of sufficient functional PGAM5 protein. Alternatively, PGAM5 might not regulate TNF-induced necroptosis in murine cells. In this regard, it is noteworthy that PGAM5 exists in two splice variants in human cells, PGAM5-L and PGAM5-S.18 Knockdown of both variants in human HT-29 cells was recently reported to protect against TNF-induced necroptosis.17 This protective phenotype could not be rescued by reintroduction of PGAM5-L alone, but required reintroduction of PGAM5-S as well.16 The fact that murine cells express PGAM5-L but not PGAM5-S might explain why a pro-necroptotic role could not be confirmed for PGAM5 in murine cells. In line with this statement is that efficient knockdown of the reported target of PGAM5, namely Drp1, caused only a small delay in TNF-necroptosis. This suggests a minor role for Drp1 and Drp1-mediated processes in TNF-induced necroptosis, such as mitochondrial fragmentation. In apparent contrast with this conclusion is the observation that addition of the Drp1 inhibitor mdivi protected against TNF-necroptosis, in line with a previous report by Wang et al.17 Overall, we conclude that at least in mouse cells the supposed Drp1 inhibitor mdvi protects from TNF-induced necroptosis by preventing respiration rather than inhibiting Drp1.
We also examined whether Pink1, a genetic interactor of PGAM520 and Drp1,21 influences necrotic cell death. Pink1 is known to affect mitochondrial fragmentation.23 In addition, Pink1 is involved in the removal of damaged mitochondria through regulation of its downstream effector Parkin,22 a ubiquitylating enzyme regulating mitophagy.25 Transient knockdown of Pink1 apparently delayed TNF-necroptosis without shifting to apoptosis, suggesting that Pink1 might be a positive regulator of TNF-necroptosis. This is an unexpected finding as Pink1 is generally reported to promote survival. Therefore, we examined the phenotype of pink1−/−MEF in TNF/Tak1i/zVAD-fmk-mediated necroptosis. Knockout of Pink1 did not affect necroptosis induction. How can this be reconciled with the apparent role of Pink1 in the first experiment? We could pinpoint the effect of the Pink1 siRNA to an off-target effect by reconstituting a mutated Pink1 that could not be knocked down. The siRNA still displayed delayed necroptosis despite the presence of non-targetable Pink1, which is in agreement with the inability of stable knockdown of Pink1 to affect TNF-necroptosis. Transient knockdown of Parkin, a downstream effector of Pink1, also did not affect TNF-necroptosis. Overall, these results indicate that processes that depend on Pink1 and Parkin, including mitophagy, are probably not important regulators of TNF-necroptosis. And although mitochondrial fission does occur during TNF-necroptosis, the absence of a protective effect after knockdown of PGAM5, Drp1, Pink1 or Parkin suggests that mitochondrial fission is probably more of a bystander phenomenon than a causal executor of TNF-necroptosis.
Altogether, our data show that not only knockdown of RIPK1, but also of the other two necrosome members RIPK3 and MLKL, blocks necroptosis and shifts the cell death modality to apoptosis, which is dependent on the kinase activity of RIPK1. None of the other supposed downstream necrosome interactors (PGAM5, Drp1) induced this switch, indicating that MLKL is the most downstream effector of necroptosis reported to induce this switch upon knockdown. It is obvious that these switches would be overlooked when studying cellular models of necroptosis that require the use of pan-caspase inhibitors or caspase depletion, as in many studies based on MEF cells. A major advantage of L929 cells is that TNF directly triggers necroptosis in the absence of pan-caspases inhibitors, whereas apoptotic signaling remains possible. We are convinced that the observed switches to apoptosis are of much importance in the search for drugs that are more specific for necroptosis by targeting RIPK3 and/or MLKL. Genetic knockdown and pharmacological inhibition do not always produce the same phenotype, especially when the proteins concerned have platform functions in addition to their enzymatic activities. Nec-1, for example, targets RIPK1 but does not induce a switch from TNF-driven necroptosis to apoptosis. Unfortunately, the recently reported inhibitor of MLKL, NSA, only targets human but not murine MLKL, and so far has been studied only in cellular models of necroptosis requiring caspase inhibition. It therefore remains to be shown in other necroptosis cellular systems, that are independent of caspase inhibitors, whether inhibition of MLKL can rescue cells from death as the cell death signal may switch to RIPK1-mediated apoptosis.
Materials and Methods
Materials
Recombinant human TNF, produced and purified to at least 99% homogeneity in our laboratory, has a specific biological activity of 3 × 107 IU/mg. Agonistic anti-human Fas (clone 2R2) was from Cell Diagnostica (Munster, Germany). zVAD-fmk was from Bachem (Bubendorf, Switzerland). Nec-1 was from Calbiochem (San Diego, CA, USA). The Tak1 inhibitor NP-009245 was purchased from AnalytiCon Discovery GmbH (Potsdam, Germany). Ac-DEVD-amc was from PeptaNova GmbH (Sandhausen, Germany). CCCP, Rotenone and Antimycin-A were purchased from Sigma Aldrich (Steinheim, Germany). NSA was from Toronto Research Chemicals (Toronto, ON, Canada). The Drp1 inhibitor mdivi was from Merck Chemicals Ltd (Nottingham, UK). Anti-β-actin (clone C4; MP BiomedicalsEurope NV, Illkirch, France), anti-Parkin (sc-32282) and anti-MLKL (sc-130172) were from Santa Cruz Biotechnology (Santa Cruz, CA, USA), anti-RIP1 from BD Biosciences (610459; Franklin Lakes, NJ, USA), and anti-RIP3 (R4277) and anti-MLKL (M6697 and SAB1408428) from Sigma Aldrich. Anti-Drp-1 was from BD Transduction laboratories (clone 8/DLP1, Franklin Lakes, NJ, USA).
Live cell imaging
Live cell imaging of L929sahFas cells stably expressing mito-GFP was performed on an AS-MDW workstation from Leica (Wetzlar, Germany). One day before imaging, 40 000 cells were seeded in a borosilicate eight-chamber plate. Next, the medium was replaced with a medium containing 50 nM TMRM and 50 μM SytoxRed (SR) for 1 h. Cells were then stimulated with TNF and fluorescence (75-W Xenon burner with monochromator set at 2 mW), and DIC images were taken every 5 min. Phototoxicity and photobleaching were prevented by minimizing the exposure time for fluorescence excitation (<100 ms) and setting the camera at gain 2 and 2 × 2 binning. Different focal planes were set at 1 μm intervals. From each image stack, maximum intensity projections (for SR, mito-GFP and TMRM) were made by using a script developed in-house for ImageJ 1.31i public domain imaging software. The 3D deconvolution (iterative restoration based on calculated PSFs) was performed on Image sequences of mito-GFP and TMRM using the Volocity software 5.2.0. Subsequent montages of the Multi-tiff time series and three-channel overlays were made in ImageJ 1.31i.
Cell culture
L929sAhFas cells were generated as previously described.2 L929sahFas with stable knockdown of Pink1 were generated by cloning short hairpins targeted against 3′UTR of Pink1 in GATEWAY compatible pcDNA6.2-GW+ EmGFP-miR (BLOCK-iT Pol II miR RNAi Expression Vector, Sigma Aldrich), according to the manufacturers instructions. Of the 10 sequences targeted in the 3′UTR in Pink1, knockdown was achieved by targeting the 3′UTR sequence cgtccagttaggttcttggg (seq 1) and accaggagttggtaagctata (seq 2) (Supplementary Data D1). pLenti6 expressing shPink1-EmGFP or mito-GFP was introduced in L929sahFas cells as described previously.29 All cell lines were cultured in DMEM supplemented with 10% fetal calf serum, penicillin (100 IU/ml), streptomycin (0.1 mg/ml), and L-glutamine (0.03%). Primary, non-immortalized MEFs were isolated and pooled from eight embryos of Pink1 KO mice or control littermates, as described previously.23
RNAi-mediated knockdown
L929sahFas cells were transfected in six-well plates according to manufacturer’s protocol as described previously.28 Knockdown was validated by RT-PCR or wherever possible, also by western blotting. Because none of the three antibodies against MLKL could reliably detect MLKL in western blots, we were compelled to validate knockdown of MLKL on the mRNA level. Reliable antibody detection is considered as clear detection of a band of expected molecular weight, of which the intensity can be reduced by knockdown, when knockdown efficiency was already confirmed by qPCR.
Cell death and caspase activity analysis
Cell death was analyzed on a FLUOSTAR Omega (BMG Labtech, Offenburg, Germany). Ten thousand cells were seeded in a transparent 96-well plate. Next day, DMEM was replaced with DMEM containing a 1.6 μM SytoxGreen (SG) and 33 μM Ac-DEVD-amc, and the cells were incubated with various inhibitors for 1 h. Next, cells were stimulated with TNF (10 000 U/ml) or agonistic anti-Fas Ab (250 ng/ml). Maximal cell death was obtained by treatment with Triton-X100 (0.05%). This allows expression of cell death as percent of control of maximal SG fluorescence (excitation 485 nm, emission 520 nm). If executor caspases-3/-7 were activated during cell death, they cleave ac-DEVD-amc only upon plasma membrane rupture. The release of fluorescent 7-amino-4-methylcoumarin (amc) was monitored (excitation 355 nm, emission 460 nm). Alternatively, cell death was analyzed on a BDPathwayTM 855 instrument (BD Biosciences) as described previously11 and by flow cytometry as previously described.29
Mitochondrial respiration
Mitochondrial oxygen consumption in the Leibovitz medium containing 1% FCS was measured in a Seahorse XF96-Analyzer (Seahorse Bioscience, Billerica, MA, USA). Oxygen consumption rates were analyzed following manufacturer’s protocols. Measurements are based on oxygen-dependent quenching of a built-in fluorescent sensor. After four measurements of basal respiration, CCCP was injected (12 μM) and maximal respiration was determined. Rotenone (2.5 μM) and Antimycin-A (5 μM) were then injected to determine the mitochondrial baseline respiration. Subsequently, the amount of cells in the microchamber of each well was determined on a BDPathwayTM 855 instrument as described previously.11 This allows normalization of the respiratory rates in function to the amount of cells in each microchamber.
Statistics
Using GraphPad Prism Version 6 (San Diego, CA, USA), data were analyzed by one-way ANOVA and a Tukey multiple comparison test, unless indicated otherwise. Statistical significance was accepted at P<0.05, number of independent experiments are mentioned and all error bars indicate standard error of the mean, unless otherwise indicated.
Abbreviations
- CCCP:
-
carbonyl cyanide m-chlorophenyl hydrazine
- cIAP:
-
cellular inhibitor of apoptosis
- DEVD-amc:
-
Asp-Glu-Val-Asp-aminoacylcoumarin
- Drp1:
-
Dynamin 1 like protein
- Mdivi:
-
mitochondrial division inhibitor
- MEF:
-
mouse embryonic fibroblast
- MLKL:
-
mixed lineage kinase domain like
- NSA:
-
necrosulfonamide
- PGAM5:
-
phosphoglycerate mutase 5
- Pink1:
-
PTEN induced putative kinase-1
- RIP:
-
receptor-interacting protein
- TMRM:
-
tetramethyl rhodamine methyl ester
- TNF:
-
tumor necrosis factor
- zVAD-fmk:
-
Z-Val-Ala-Asp-fluoromethylketone
References
Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G . Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol 2010; 11: 700–714.
Vercammen D, Beyaert R, Denecker G, Goossens V, Van Loo G, Declercq W et al. Inhibition of caspases increases the sensitivity of L929 cells to necrosis mediated by tumor necrosis factor. J Exp Med 1998; 187: 1477–1485.
Degterev A, Hitomi J, Germscheid M, Ch'en IL, Korkina O, Teng X et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol 2008; 4: 313–321.
Newton K, Sun X, Dixit VM . Kinase RIP3 is dispensable for normal NF-kappa Bs, signaling by the B-cell and T-cell receptors, tumor necrosis factor receptor 1 and Toll-like receptors 2 and 4. Mol Cell Biol 2004; 24: 1464–1469.
Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol 2005; 1: 112–119.
Linkermann A, Bräsen J, Himmerkus N, Liu S, Huber T, Kunzendorf U et al. Rip1 (receptor-interacting protein kinase 1) mediates necroptosis and contributes to renal ischemia/reperfusion injury. Kidney Int 2012; 81: 751–761.
Gunther C, Martini E, Wittkopf N, Amann K, Weigmann B, Neumann H et al. Caspase-8 regulates TNF-alpha-induced epithelial necroptosis and terminal ileitis. Nature 2011; 477: 335–339.
Welz PS, Wullaert A, Vlantis K, Kondylis V, Fernandez-Majada V, Ermolaeva M et al. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature 2011; 477: 330–334.
Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 2009; 137: 1112–1123.
Robinson N, McComb S, Mulligan R, Dudani R, Krishnan L, Sad S . Type I interferon induces necroptosis in macrophages during infection with Salmonella enterica serovar Typhimurium. Nat Immunol 2012; 13: 954–962.
Duprez L, Takahashi N, Van Hauwermeiren F, Vandendriessche B, Goossens V, Vanden Berghe T et al. RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity 2011; 35: 908–918.
He S, Wang L, Miao L, Wang T, Du F, Zhao L et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell 2009; 137: 1100–1111.
Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 2009; 325: 332–336.
Vanlangenakker N, Bertrand M, Bogaert P, Vandenabeele P, Vanden Berghe T . TNF-induced necroptosis in L929 cells is tightly regulated by multiple TNFR1 complex I and II members. Cell Death Dis 2011; 2: e230.
Sun L, Wang H, Wang Z, He S, Chen S, Liao D et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 2012; 148: 213–227.
Zhao J, Jitkaew S, Cai Z, Choksi S, Li Q, Luo J et al. Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc Natl Acad Sci USA 2012; 109: 5322–5327.
Wang Z, Jiang H, Chen S, Du F, Wang X . The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 2012; 148: 228–243.
Lo S-C, Hannink M . PGAM5 a Bcl-XL-interacting protein, is a novel substrate for the redox-regulated Keap1-dependent ubiquitin ligase complex. J Biol Chem 2006; 281: 37893–37903.
Takeda K, Komuro Y, Hayakawa T, Oguchi H, Ishida Y, Murakami S et al. Mitochondrial phosphoglycerate mutase 5 uses alternate catalytic activity as a protein serine/threonine phosphatase to activate ASK1. Proc Natl Acad Sci USA 2009; 106: 12301–12305.
Imai Y, Kanao T, Sawada T, Kobayashi Y, Moriwaki Y, Ishida Y et al. The loss of PGAM5 suppresses the mitochondrial degeneration caused by inactivation of PINK1 in Drosophila. PLoS Genet 2010; 6: 12.
Lutz A, Exner N, Fett M, Schlehe J, Kloos K, Lämmermann K et al. Loss of parkin or PINK1 function increases Drp1-dependent mitochondrial fragmentation. J Biol Chem 2009; 284: 22938–22951.
Arena G, Gelmetti V, Torosantucci L, Vignone D, Lamorte G, De Rosa P et al. PINK1 protects against cell death induced by mitochondrial depolarization, by phosphorylating Bcl-xL and impairing its pro-apoptotic cleavage. Cell Death Differ 2013; 20: 920–930.
Heeman B, Van den Haute C, Aelvoet S-A, Valsecchi F, Rodenburg R, Reumers V et al. Depletion of PINK1 affects mitochondrial metabolism, calcium homeostasis and energy maintenance. J Cell Sci 2011; 124 (Pt 7): 1115–1125.
Youle R, Narendra D . Mechanisms of mitophagy. Nat Rev Mol Cell Biol 2011; 12: 9–14.
Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier C et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. The J Cell Biol 2010; 189: 211–221.
Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ et al. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 2008; 135: 1311–1323.
Vanden Berghe T, Vanlangenakker N, Parthoens E, Deckers W, Devos M, Festjens N et al. Necroptosis, necrosis and secondary necrosis converge on similar cellular disintegration features. Cell Death Differ 2010; 17: 922–930.
Festjens N, Kalai M, Smet J, Meeus A, Van Coster R, Saelens X et al. Butylated hydroxyanisole is more than a reactive oxygen species scavenger. Cell Death Differ 2006; 13: 166–169.
Vanlangenakker N, Vanden Berghe T, Bogaert P, Laukens B, Zobel K, Deshayes K et al. cIAP1 and TAK1 protect cells from TNF-induced necrosis by preventing RIP1/RIP3-dependent reactive oxygen species production. Cell Death Differ 2011; 18: 656–665.
Cassidy-Stone A, Chipuk J, Ingerman E, Song C, Yoo C, Kuwana T et al. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell 2008; 14: 193–204.
Declercq W, Vanden Berghe T, Vandenabeele P . RIP kinases at the crossroads of cell death and survival. Cell 2009; 138: 229–232.
Pobezinskaya YL, Kim YS, Choksi S, Morgan MJ, Li T, Liu C et al. The function of TRADD in signaling through tumor necrosis factor receptor 1 and TRIF-dependent Toll-like receptors. Nat Immunol 2008; 9: 1047–1054.
Ermolaeva MA, Michallet MC, Papadopoulou N, Utermohlen O, Kranidioti K, Kollias G et al. Function of TRADD in tumor necrosis factor receptor 1 signaling and in TRIF-dependent inflammatory responses. Nat Immunol 2008; 9: 1037–1046.
Schneider-Brachert W, Tchikov V, Neumeyer J, Jakob M, Winoto-Morbach S, Held-Feindt J et al. Compartmentalization of TNF receptor 1 signaling: internalized TNF receptosomes as death signaling vesicles. Immunity 2004; 21: 415–428.
Wilson NS, Dixit V, Ashkenazi A . Death receptor signal transducers: nodes of coordination in immune signaling networks. Nat Immunol 2009; 10: 348–355.
Wang L, Du F, Wang X . TNF-alpha induces two distinct caspase-8 activation pathways. Cell 2008; 133: 693–703.
Biton S, Ashkenazi A . NEMO and RIP1 Control Cell Fate in Response to Extensive DNA Damage via TNF-alpha Feedforward Signaling. Cell 2011; 145: 92–103.
Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F et al. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol Cell 2011; 43: 432–448.
Feoktistova M, Geserick P, Kellert B, Dimitrova D, Langlais C, Hupe M et al. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell 2011; 43: 449–463.
Dondelinger Y, Aguileta M, Goossens V, Dubuisson C, Grootjans S, Dejardin E et al. RIPK3 contributes to TNFR1-mediated RIPK1 kinase-dependent apoptosis in conditions of cIAP1/2 depletion or TAK1 kinase inhibition. Cell Death Differ 2013; 20: 1381–1392.
Murphy J, Czabotar P, Hildebrand J, Lucet I, Zhang J-G, Alvarez-Diaz S et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity 2013; 39: 443–453.
Acknowledgements
We thank Amin Bredan for editing the manuscript. PV is senior full professor at Ghent University and holder of a Methusalem grant. TV and QR hold a postdoctoral researcher position with the FWO (Fund for Scientific Research, Flanders). YD is holder of a PhD fellowship from the Agency for Innovation by Science and Technology. MB has a tenure track position in the Multidisciplinary Research Program of Ghent University. NT, SG and VG are paid from the Methusalem grant. Research in the Vandenabeele group is supported by Belgian grants (Interuniversity Attraction Poles, IAP 7/32), Flemish grants (Research Foundation Flanders – FWO G.0875.11, FWO G.0973.11, FWO G.0A45.12N, FWO G.0787.13N Methusalem grant – BOF09/01M00709), Ghent University grants (MRP, GROUP-ID consortium), grant from the Foundation against Cancer, (F94) and grants from the VIB.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Melino
Supplementary Information accompanies this paper on Cell Death and Disease website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Remijsen, Q., Goossens, V., Grootjans, S. et al. Depletion of RIPK3 or MLKL blocks TNF-driven necroptosis and switches towards a delayed RIPK1 kinase-dependent apoptosis. Cell Death Dis 5, e1004 (2014). https://doi.org/10.1038/cddis.2013.531
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.531
Keywords
This article is cited by
-
Necrostatin-1S mitigates type-2 diabetes-associated cognitive decrement and lipotoxicity-induced neuro-microglia changes through p-RIPK-RIPK3-p-MLKL axis
Metabolic Brain Disease (2023)
-
The development of necroptosis: what we can learn
Cell Stress and Chaperones (2023)
-
RIP1-dependent linear and nonlinear recruitments of caspase-8 and RIP3 respectively to necrosome specify distinct cell death outcomes
Protein & Cell (2021)
-
Gut stem cell necroptosis by genome instability triggers bowel inflammation
Nature (2020)
-
Cyclin-Dependent Kinase 9 Inhibition Suppresses Necroptosis and Pyroptosis in the Progress of Endotoxemia
Inflammation (2020)