Abstract
Smac mimetic promotes apoptosis by neutralizing inhibitor of apoptosis (IAP) proteins and is considered as a promising cancer therapeutic. Although an autocrine/paracrine tumor necrosis factor-α (TNFα) loop has been implicated in Smac mimetic-induced cell death, little is yet known about additional factors that determine sensitivity to Smac mimetic. Using genome-wide gene expression analysis, we identify death receptor 5 (DR5) as a novel key mediator of Smac mimetic-induced apoptosis. Although several cell lines that are sensitive to the Smac mimetic BV6 die in a TNFα-dependent manner, A172 glioblastoma cells undergo BV6-induced apoptosis largely independently of TNFα/TNFR1, as the TNFα-blocking antibody Enbrel or TNFR1 knockdown provide little protection. Yet, BV6-stimulated nuclear factor-κB (NF-κB) activation is critically required for apoptosis, as inhibition of NF-κB by overexpression of dominant-negative IκBα superrepressor (IκBα-SR) blocks BV6-induced apoptosis. Unbiased genome-wide gene expression studies in IκBα-SR-overexpressing cells versus vector control cells reveal that BV6 increases DR5 expression in a NF-κB-dependent manner. Importantly, this BV6-stimulated upregulation of DR5 is critically required for apoptosis, as transient or stable knockdown of DR5 significantly inhibits BV6-triggered apoptosis. In addition, DR5 silencing attenuates formation of a RIP1/FADD/caspase-8 cytosolic cell death complex and activation of caspase-8, -3 and -9. By identifying DR5 as a critical mediator of Smac mimetic-induced apoptosis, our findings provide novel insights into the determinants that control susceptibility of cancer cells to Smac mimetic.
Similar content being viewed by others
Main
Cell death by apoptosis represents a fundamental cellular program that is critically involved in the regulation of various physiological processes.1 Human cancers are characterized by their inability to properly respond to apoptotic stimuli, as apoptosis is typically impaired during tumorigenesis.2 As implied by their name, membrane-bound death receptors of the tumor necrosis factor (TNF) receptor 1 (TNFR1) superfamily represent prototypic entry points for external signals to initiate apoptosis and can also sense intracellular cues.3 In the death receptor (extrinsic) pathway, the binding of death receptors such as TNF-related apoptosis-inducing ligand receptors (TRAIL receptors) by their natural ligands or agonistic antibodies leads to activation of caspase-8 in a receptor-associated death-inducing signaling complex (DISC) that mediates apoptosis by cleaving and activating downstream effector caspases such as caspase-3.3 Furthermore, internalization of ligand-bound death receptors can subsequently lead to the assembly of a multimeric cell death complex in the cytosol known as complex II that contains receptor-interacting protein 1 (RIP1), FAS-associated via death domain (FADD) and caspase-8.4
Apoptosis pathways are tightly regulated by pro- and antiapoptotic proteins.2 For example, ‘Inhibitor of Apoptosis’ (IAP) proteins represent a family of antiapoptotic proteins with highly expressed levels in many human cancers.5 X-linked inhibitor of apoptosis protein (XIAP) inhibits caspase activation by binding caspase-9 and -3/-7 via its baculovirus IAP repeat (BIR) domains.6 IAP proteins harboring a RING (really interesting new gene) domain, for example, cellular inhibitor of apoptosis (cIAP)1 and cIAP2, can act as E3 ubiquitin ligases and mediate auto- and heteroubiquitination and subsequent proteasomal degradation.5 IAP proteins are known as key regulators of canonical and non-canonical nuclear factor-κB (NF-κB) pathways.5 In the canonical NF-κB pathway, IκBα gets phosphorylated and degraded upon TNFR1 activation, leading to p65/p50 nuclear translocation and transcriptional activation of NF-κB target genes.7 cIAP proteins promote activation of the canonical NF-κB pathway by non-degradative ubiquitination of the serine/threonine kinase RIP1.8, 9 In the non-canonical NF-κB pathway, cIAP proteins constitutively stimulate proteasomal degradation of NF-κB-inducing kinase (NIK) in a multiprotein complex together with TNF receptor-associated factor 2 (TRAF2) and TRAF3, thereby preventing proteolytic processing of p100 to p52 and nuclear translocation of p52.10, 11
IAP proteins are held in check by endogenous antagonists including second mitochondria-derived activator of caspase (Smac).5 Upon its release from the mitochondria into the cytosol, Smac triggers caspase-3 activation by neutralizing XIAP.5 Small-molecule pharmacological inhibitors of IAP proteins such as Smac mimetic have been developed that antagonize XIAP and also stimulate E3 ligase activity of IAP proteins with a RING domain such as cIAP1/2 and XIAP, leading to their autoubiquitination and proteasomal degradation.12, 13, 14 Depletion of cIAP1/2 by Smac mimetic favors deubiquitination of RIP1 that, in turn, facilitates TNFα-induced cell death by enhancing the formation of complex II in the cytosol that contains RIP1/FADD/caspase-8.8, 9 In addition, downregulation of cIAP1/2 leads to activation of NF-κB signaling via accumulation of NIK that, in turn, initiates a TNFα/TNFR1 autocrine/paracrine loop that has been reported to mediate Smac mimetic-induced cytotoxicity.12, 13 Accordingly, sensitivity of cancer cells to Smac mimetic has been linked to their ability to activate TNFα/TNFR1 autocrine/paracrine signaling. However, little is yet known about the molecular determinants that are critical for sensitivity of cancer cells to Smac mimetic. Therefore, in this study we aimed to identify novel regulators of Smac mimetic-induced apoptosis.
Results
TNFα/TNFR1 signaling is largely dispensable for BV6-mediated apoptosis in A172 glioblastoma cells
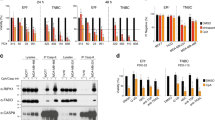
To elucidate the molecular mechanisms of BV6-induced cell death, we analyzed the effects of BV6 in cell lines of different cancer entities. Treatment with BV6 resulted in a dose-dependent reduction of cell viability in A172 glioblastoma cells, MDA-MB-231 breast carcinoma cells, SK-N-AS neuroblastoma cells and Kym-1 rhabdomyosarcoma cells (Figure 1a and Supplementary Figure 1a). In addition, BV6 triggered DNA fragmentation, a biochemical hallmark of apoptosis, in a concentration-dependent manner (Figure 1b and Supplementary Figure 1b), demonstrating that BV6 induces apoptotic cell death. Furthermore, the broad-range caspase inhibitor Z-Val-Ala-Asp (OMe)-fluoromethylketone (zVAD.fmk) significantly reduced BV6-induced loss of cell viability as well as DNA fragmentation (Figures 1a and b), in line with caspase-dependent apoptosis.
BV6-mediated apoptosis in A172 cells is mainly TNFα-independent. (a and b) Cells were treated for 72 h with indicated concentrations of BV6 in the presence or absence of 20 μM zVAD.fmk or 10 μg/ml Enbrel. Cell viability was measured by MTT or crystal violet assay and is expressed as the percentage of untreated controls (a). Apoptosis was determined by DNA fragmentation of PI-stained nuclei using flow cytometry (b). Mean±S.D. of three independent experiments performed in duplicate are shown. (c–e) Cells were transiently transfected with siRNA against TNFR1 or control siRNA and treated for 72 h with indicated concentrations of BV6. Protein expression of TNFR1 was analyzed by western blotting, and GAPDH served as loading control (c). Cell viability was measured by MTT assay and is expressed as the percentage of untreated controls (d). Apoptosis was determined by DNA fragmentation of PI-stained nuclei using flow cytometry and the percentage of specific apoptosis is shown (e). Mean±S.E.M. of three to four independent experiments performed in duplicate are shown
As a TNFα-driven autocrine/paracrine loop that is initiated upon depletion of cIAP proteins has been implicated to mediate Smac mimetic-induced cell death,12, 13, 14 we next explored the involvement of TNFα/TNFR1 signaling. To this end, we pharmacologically interfered with TNFα/TNFR1 signaling by using the TNFα-blocking antibody Enbrel. Unexpectedly, Enbrel largely failed to rescue BV6-induced cell death in A172 cells, as it did not prevent BV6-triggered reduction of cell viability and DNA fragmentation (Figures 1a and b and Supplementary Figure 2). In contrast, Enbrel significantly inhibited BV6-induced loss of cell viability and DNA fragmentation in MDA-MB-231, SK-N-AS and Kym1 cells (Figures 1a and b).
In addition to this pharmacological approach, we also employed a genetic strategy to block TNFα/TNFR1 signaling using RNAi-mediated knockdown of TNFR1. We selected A172 and MDA-MB-231 as prototypic cell lines that were (MDA-MB-231 cells) or were not (A172 cells) protected by Enbrel against BV6-induced apoptosis. Similarly, TNFR1 silencing largely failed to protect against Smac mimetic-induced loss of cell viability and DNA fragmentation in A172 cells (Figures 1c–e). Control experiments showed that TNFR1 silencing suppressed TNFα-stimulated signaling in A172 cells, as assessed by inhibition of IκBα phosphorylation (Supplementary Figure 3). In contrast, TNFR1 knockdown significantly inhibited Smac mimetic-induced cell death in MDA-MB-231 cells (Figures 1c–e). These data point to a cell line-dependent requirement of TNFα/TNFR1 signaling in Smac mimetic-induced cell death. To further investigate the underlying molecular mechanisms, we used A172 and MDA-MB-231 as prototypic cell lines that undergo apoptosis upon treatment with BV6 in a TNFα-independent and TNFα-dependent manner, respectively.
Depletion of IAP proteins and activation of caspases by BV6
As Smac mimetic has been shown to stimulate ubiquitination and proteasomal degradation of IAP proteins harboring a RING domain,12, 13, 14 we next examined IAP protein levels upon exposure to BV6. Treatment with BV6 caused downregulation of cIAP1 and XIAP in both A172 and MDA-MB-231 cells (Figure 2a). Furthermore, depletion of cIAP proteins by Smac mimetic has been reported to increase formation of a cytosolic multimeric complex comprising RIP1, FADD and caspase-8, as cIAP proteins can act as E3 ligases for ubiquitination of RIP1.12, 13, 14 Indeed, immunoprecipitation of caspase-8 revealed that BV6 stimulated the interaction of RIP1, FADD and caspase-8 in a time-dependent manner (Figure 2b). As this cytosolic RIP1/FADD/caspase-8-containing complex has been described to drive caspase-8 activation,8, 9 we next monitored activation of the caspase cascade by western blotting. BV6 induced processing of caspase-8, -9 and -3 into active cleavage fragments as indicated by the appearance of caspase-8 cleavage products p43/41 and p18, caspase-9 cleavage fragments p37/p35 and caspase-3 cleavage products p17/12 (Figure 2c). Cleavage of caspases was accompanied by decreased expression levels of the proenzyme forms of caspase-8, -9 and -3, especially in MDA-MB-231 cells (Figure 2c), confirming that caspases are activated by proteolytic cleavage.
BV6 triggers depletion of IAP proteins and caspase activation. (a) A172 and MDA-MB-231 cells were treated for indicated times with 5 μM BV6 (A172) or 50 nM BV6 (MDA-MB-231). Expression levels of cIAP1 and XIAP were analyzed by western blotting. Expression of GAPDH served as loading control. (b) A172 cells were treated for indicated times with 5 μM BV6 in the presence of 20 μM zVAD.fmk. Caspase-8 was immunoprecipitated using an anti-caspase-8 antibody and detection of indicated proteins was done by western blotting. (c) A172 and MDA-MB-231 cells were treated for indicated times with 5 μM BV6 (A172) or 50 nM BV6 (MDA-MB-231); asterisks indicate unspecific bands. Caspase activation was analyzed by western blotting, and active cleavage fragments are indicated by arrow heads. Expression of β-actin and GAPDH served as loading controls
BV6 activates canonical and non-canonical NF-κB signaling
As Smac mimetic-mediated apoptosis has been reported to engage TNFα/TNFR1 signaling by activating NF-κB upon depletion of IAP proteins,12, 13, 14 we asked whether the observed differential requirement of TNFα/TNFR1 signaling is due to differences in BV6-stimulated NF-κB activation. To assess non-canonical NF-κB activation, we determined accumulation of NIK protein and proteolytic processing of p100 to p52. For canonical NF-κB activation, we analyzed phosphorylation of IκBα and p65 as well as degradation of IκBα. BV6 caused accumulation of NIK and processing of p100 to p52 in both A172 and MDA-MB-231 cells (Figure 3a). In addition, BV6 stimulated phosphorylation of p65 and IκBα that was accompanied by downregulation of IκBα protein levels in both A172 and MDA-MB-231 cells (Figure 3a). As positive control for canonical NF-κB signaling, we used TNFα that potently stimulated phosphorylation and degradation of IκBα protein, whereas it did not cause NIK accumulation or p100 processing (Figure 3a). The more pronounced degradation of IκBa in TNFα- versus BV6-treated cells may be due to stronger and more rapid NF-κB activation by TNFα.
BV6 activates the canonical and non-canonical NF-κB pathway. (a) A172 and MDA-MB-231 cells were treated for indicated times with 5 μM BV6 (A172) or 50 nM BV6 (MDA-MB-231); stimulation with 10 ng/ml TNFα was used as prototypic canonical NF-κB stimulus. Expression and/or phosphorylation of NIK, p100, p52, p65 and IκBα were analyzed by western blotting. Expression of GAPDH served as loading control. (b) A172 and MDA-MB-231 cells were treated for indicated times with 5 μM BV6 (A172) or 50 nM BV6 (MDA-MB-231). Expression levels of p50, p52, p65 and RelB were analyzed in the nucleus or in the cytosol by western blotting. α-tubulin served as purity control for cytoplasmic fractions, lamin B1 for nuclear fractions and β-actin as loading control for both fractions. (c) A172 and MDA-MB-231 cells were treated for 24 h with 5 μM BV6 (A172) or 50 nM BV6 (MDA-MB-231). NF-κB transcriptional activity was determined using a FITC-labeled NF-κB reporter construct. Fold increase in NF-κB transcriptional activity is shown with mean±S.D. of three independent experiments performed in at least triplicate. (d) A172 and MDA-MB-231 cells were treated for indicated times with 5 μM BV6 (A172) or 50 nM BV6 (MDA-MB-231). mRNA expression levels of TNFα, p100 and RelB were analyzed by RT-qPCR. Fold increase in RNA levels is shown with mean+S.E.M. of two to five independent experiments performed in triplicate
To analyze which NF-κB subunits translocate into the nucleus, we prepared cytosolic and nuclear extracts. BV6 stimulated nuclear translocation of p52, p50, RelB and phospho-p65 in both A172 and MDA-MB-231 cells (Figure 3b). Furthermore, analysis of DNA-binding by EMSA showed that BV6 increased NF-κB DNA binding (Supplementary Figure 4). To determine whether NF-κB DNA binding leads to transcriptional activation of NF-κB target genes, we examined both NF-κB transcriptional activity utilizing a NF-κB-driven reporter construct and NF-κB target gene expression using reverse transcriptase quantitative PCR (RT-qPCR) analysis. BV6 significantly increased NF-κB transcriptional activity (Figure 3c) and upregulated prototypic NF-κB target genes including TNFα, RelB and p100 in both A172 and MDA-MB-231 cells (Figure 3d). These experiments showed that BV6 activates canonical and non-canonical NF-κB signaling in A172 and MDA-MB-231 cells.
NF-κB is necessary for BV6-induced cell death
We next asked whether NF-κB activation is required for BV6-induced cell death. To block NF-κB activation we overexpressed IκBα superrepressor (IκBα-SR) (Figure 4a). Control experiments showed that IκBα overexpression suppressed BV6-stimulated phosphorylation of IκBα and protein levels of NF-κB target genes such as IκBα, p100 and RelB (Figure 4b). In addition, IκBα-SR inhibited NF-κB DNA binding and reduced mRNA levels of NF-κB target genes, that is, TNFα, p100 and RelB (Supplementary Figure 5), indicating that IκBα-SR suppresses both canonical and non-canonical NF-κB signaling. Importantly, inhibition of NF-κB by IκBα-SR significantly reduced BV6-stimulated loss of cell viability and DNA fragmentation compared with vector control cells in both A172 and MDA-MB-231 cells (Figures 4c and d). In addition, IκBα-SR overexpression profoundly inhibited caspase activation and attenuated the formation of the RIP1/FADD/caspase-8 cytosolic cell death complex upon treatment with BV6 compared with vector control cells (Figures 4e and f). Together, this set of experiments demonstrates that NF-κB is required for BV6-stimulated cell death in A172 and MDA-MB-231 cells.
NF-κB is required for BV6-induced cell death. (a) A172 and MDA-MB-231 cells were transduced with IκBα-SR or vector control. Expression of IκBα and IκBα-SR was analyzed by western blotting. GAPDH expression served as loading control. (b) A172 and MDA-MB-231 cells were transduced with IκBα-SR or vector control. Cells were treated for indicated times with 5 μM BV6 (A172) or 50 nM BV6 (MDA-MB-231). Expression and/or phosphorylation of cIAP1, p100, p52, RelB and IκBα were analyzed by western blotting. Expression of GAPDH served as loading control. (c and d) A172 and MDA-MB-231 cells stably expressing IκBα-SR or vector control were treated for 72 h with indicated concentrations of BV6. Cell viability was measured by MTT or crystal violet assay and is expressed as the percentage of untreated controls (c). Apoptosis was determined by DNA fragmentation of PI-stained nuclei using flow cytometry (d). Mean±S.D. of three to seven independent experiments performed in duplicate are shown. (e) A172 cells stably expressing IκBα-SR or vector control were treated for indicated times with 5 μM BV6. Caspase activation was analyzed by western blotting, and active cleavage fragments are indicated by arrow heads; β-actin served as loading control; asterisks indicate unspecific bands. (f) A172 cells were treated for 24 h with 5 μM BV6 in the presence of 20 μM zVAD.fmk. Caspase-8 was immunoprecipitated using an anti-caspase-8 antibody and detection of indicated proteins was done by western blotting
Identification of DR5 as an NF-κB target gene that is upregulated by BV6
As in this study TNFα turned out to be largely dispensable for BV6-induced cell death in A172 cells despite the requirement of NF-κB, we next aimed to identify NF-κB target genes other than TNF α that act as critical mediators of BV6-induced apoptosis. To this end, we performed a genome-wide cDNA microarray analysis and compared BV6-stimulated gene expression in A172 cells overexpressing IκBα-SR with vector control cells. Gene set enrichment analysis (GSEA) verified strong enrichment of NF-κB target genes under treated NF-κB-proficient conditions, proving functionality of IκBα-SR expression (Supplementary Table 2). It is noteworthy that LRIG1, which has already been described in the context of transcriptional regulation by Smac mimetic,15 was displayed among the top hits and has been validated to be regulated by BV6 treatment in a NF-κB-dependent manner (Supplementary Figure 6). Interestingly, by this approach we identified death receptor 5 (DR5), another death receptor of the TNFR superfamily, as one of the top listed NF-κB-regulated genes that is differentially upregulated upon treatment with BV6 in A172 vector control compared with IκBα-SR-overexpressing cells. Validation experiments using RT-qPCR and western blot analysis confirmed that treatment with BV6 triggered upregulation of DR5 mRNA and protein expression (Figures 5a and b). To ensure that DR5 is regulated in a NF-κB-dependent manner, we compared DR5 expression in IκBα-SR-overexpressing A172 cells with vector control cells. It is noteworthy that IκBα-SR overexpression prevented the BV6-stimulated increase in DR5 mRNA and protein expression (Figures 5c and d). In addition, constitutive expression of DR5 mRNA and protein was reduced in IκBα-SR-overexpressing cells compared with vector control cells (Figures 5c and d). These findings demonstrate that BV6 triggers upregulation of DR5 in a NF-κB-dependent manner.
BV6 stimulates upregulation of DR5 via NF-κB. (a and b) A172 cells were treated for indicated times with 5 μM BV6. DR5 mRNA levels were analyzed by RT-qPCR and fold increase in DR5 mRNA levels is shown (a). Mean±S.E.M. values of three independent experiments are shown. DR5 protein expression was analyzed by western blotting; β-actin served as loading control (b). (c and d) A172 cells stably expressing IκBα-SR or vector control were treated for indicated times with 5 μM BV6. DR5 mRNA levels were analyzed by RT-qPCR and fold increase in DR5 mRNA levels is shown (c). Mean±S.D. values of five independent experiments are shown. DR5 protein expression was analyzed by western blotting; β-actin served as loading control (d)
DR5 upregulation is critically required for BV6-mediated cell death
To investigate whether BV6-stimulated upregulation of DR5 is necessary for BV6-mediated cell death, we knocked down DR5 by siRNA. Control experiments confirmed that two distinct siRNA sequences targeting DR5 profoundly suppressed constitutive expression and BV6-stimulated upregulation of DR5 mRNA and protein (Figures 6a and b). Importantly, knockdown of DR5 strongly inhibited BV6-triggered loss of viability and DNA fragmentation (Figures 6c and d). In addition, the BV6-induced cleavage of caspase-8, -9 and -3 into active fragments and formation of the RIP1/FADD/caspase-8 complex was severely impaired in DR5 knockdown cells compared with control cells (Figures 6e and f). Interestingly, the caspase-8 cleavage products p43 and p41 were detected upon BV6 treatment within the cytosolic RIP1/FADD/caspase-8 complex in control cells, but not in DR5 knockdown cells (Figure 6f), underlining that DR5-mediated formation of this complex is required to drive caspase-8 activation.
DR5 upregulation is necessary for BV6-mediated cell death. A172 cells were transiently transfected with 5 nM siRNA targeting DR5 or control siRNA and treated for 6 h (RT-qPCR) or 24 h (western blotting) with 5 μM BV6. Protein expression of DR5 was analyzed by western blotting, and GAPDH served as loading control (a). DR5 mRNA levels were analyzed by RT-qPCR and fold increase in DR5 mRNA levels is shown (b). Mean±S.D. values of two independent experiments are shown. Cell viability was measured by MTT assay and is expressed as the percentage of untreated controls (c). Apoptosis was determined by DNA fragmentation of PI-stained nuclei using flow cytometry and the percentage of specific apoptosis is shown (d). Mean±S.D. of three to four independent experiments performed in duplicate are shown. Caspase activation was analyzed by western blotting, active cleavage fragments are indicated by arrow heads; β-actin served as loading control; asterisks indicate unspecific bands (e). Caspase-8 was immunoprecipitated using an anti-caspase-8 antibody and detection of indicated proteins was done by western blotting (f)
In a second genetic approach to block DR5, we used three distinct short-hairpin RNA (shRNA) vectors that prevented in particular the BV6-stimulated DR5 upregulation rather than constitutive expression of DR5 (Supplementary Figure 7a). Abrogation of BV6-mediated upregulation of DR5 significantly inhibited BV6-induced loss of viability and DNA fragmentation in DR5 knockdown cells compared with control cells (Supplementary Figures 7b and c). Together, these independent approaches to block DR5 demonstrate that DR5 is necessary for BV6-mediated formation of the RIP1/FADD/caspase-8 complex, caspase activation and cell death in A172 cells. In comparison, silencing of DR5 failed to rescue MDA-MB-231 cells from BV6-induced apoptosis, although BV6 triggered a modest increase of DR5 mRNA and protein levels in these cells (Supplementary Figures 8a–d). Control experiments showed that DR5 silencing suppressed ETR2-induced cell death in MDA-MB-231 cells, verifying a functional knockdown (Supplementary Figure 8e).
As TRAIL is known as another NF-κB target gene, we explored the possibility that TRAIL is involved in mediating BV6-induced apoptosis. BV6 slightly increased TRAIL mRNA expression in a time-dependent manner (Supplementary Figure 9a). The addition of a TRAIL-blocking antibody failed to protect against BV6-triggered loss of viability under conditions where it fully prevented TRAIL-induced cell death (Supplementary Figure 9b). In comparison, genetic silencing of TRAIL provided some protection against BV6-induced loss of viability at low doses of BV6 (Supplementary Figures 9c and d). These results suggest that TRAIL may directly bind to DR5 as transmembrane protein and is therefore spared from the inhibitory actions of the TRAIL-blocking antibody. Together, this set of experiments indicates that BV6-induced apoptosis in A172 cells is mediated via DR5 in a partially TRAIL-dependent manner.
Discussion
Smac mimetic is considered as a promising cancer therapeutic to initiate cell death in cancer cells. Although an autocrine/paracrine TNFα loop has been implicated to mediate Smac mimetic-induced cell death, little is yet known about additional factors that determine sensitivity of cancer cells to Smac mimetic-triggered apoptosis. Using an unbiased genome-wide gene expression profiling approach, we identify DR5 as a novel key mediator of Smac mimetic-induced apoptosis. Several lines of evidence support this conclusion (Figure 7). First, BV6-triggered increase in DR5 mRNA and protein expression is critically required for BV6-induced apoptosis, as knockdown of DR5 using transient and stable strategies for gene silencing strongly reduces apoptosis by BV6. DR5 initiates apoptosis by promoting the formation of a RIP1/FADD/caspase-8 cell death complex in the cytosol that drives activation of caspase-8, -9 and -3 and apoptosis, as all these events are inhibited by DR5 silencing. DR5 mediates BV6-induced apoptosis in a soluble ligand-independent manner, as a TRAIL-blocking antibody fails to rescue BV6-induced apoptosis under conditions where it blocks TRAIL-induced cell death. Second, TNFα/TNFR1 signaling is largely dispensable for BV6-induced apoptosis in a cell type-dependent manner, as pharmacological or genetic inhibition of the TNFα/TNFR1 ligand/receptor system affords little protection of A172 glioblastoma cells against BV6-induced apoptosis. Third, BV6 stimulates DR5 expression in a NF-κB-dependent manner, as inhibition of NF-κB by overexpression of dominant-negative IκBα-SR abolishes DR5 upregulation by BV6. Together, these findings demonstrate that – besides an autocrine/paracrine TNFα/TNFR1 loop – BV6-stimulated activation of DR5 can serve as an entry site to engage the apoptotic machinery upon treatment with BV6.
Scheme of BV6-induced signaling pathways. BV6 triggers NF-κB activation and induction of target genes such as TNFα or DR5. NF-κB mediates BV6-induced apoptosis in a TNFα-dependent manner via an autocrine/paracrine TNFα/TNFR1 loop or, alternatively, in a DR5-dependent manner. BV6-stimulated, NF-κB-dependent upregulation of DR5 is required for the formation of a cytosolic complex consisting of caspase-8, RIP1 and FADD, subsequent caspase activation and apoptosis
The novelty of our study particularly resides in the discovery of DR5 as a key mediator of Smac mimetic-induced apoptosis. So far, upregulation of DR5 in response to treatment with Smac mimetic has not yet been reported. DR5 is known as a NF-κB target gene that harbors a NF-κB consensus binding site within its first intronic region.16, 17, 18 Consistently, we found that BV6 increases DR5 mRNA and protein expression in a NF-κB-dependent manner. DR5 is required for the formation of a cytosolic cell death complex composed of RIP1, FADD and caspase-8 upon treatment with BV6 that drives caspase-8 activation. In support of this notion, knockdown of DR5 abolishes cleavage of caspase-8 within the cytosolic RIP1/FADD/caspase-8 complex. It is interesting to note that this RIP1/FADD/ caspase-8-containing cytosolic complex has been reported to assemble upon treatment with Smac mimetic in a TNFα-dependent manner.12, 13, 19 This indicates that Smac mimetic activates death receptor/ligands systems, that is, TNFα/TNFR1 or DR5, via NF-κB in a context-dependent manner, which in turn promote the assembly of the RIP1/FADD/caspase-8-containing complex in the cytosol that drives caspase-8 activation and apoptosis.
Our study indicates that DR5 mediates BV6-induced apoptosis in a partially TRAIL-dependent manner independently of the soluble form of the ligand, as genetic silencing of TRAIL partially rescues cell death at low concentrations of BV6, whereas pharmacological inhibition of TRAIL fails to protect against BV6-induced apoptosis. Although previous studies investigated the contribution of the ligand TRAIL to Smac mimetic-induced apoptosis and demonstrated that knockdown of TRAIL, TRAIL-blocking antibodies or TRAIL receptor decoy constructs that sequester TRAIL were all unable to afford protection against Smac mimetic-induced apoptosis,12, 13, 19 our present study particularly addressed the role of TRAIL receptors. Of the two proapoptotic TRAIL receptors, DR5 is especially relevant for glioblastoma, as DR4 is frequently epigenetically silenced in this tumor.20
Previously, sensitivity to Smac mimetic has been linked to activation of TNFα/TNFR1, another ligand/receptor system of the death receptor family. Accordingly, Smac mimetic-triggered depletion of cIAP proteins was shown to initiate a TNFα/TNFR1 autocrine/paracrine loop via NIK accumulation and NF-κB activation.12, 13 Consistent with these reports,12, 13 we found that several BV6-sensitive cell lines die via a TNFα/TNFR1 autocrine/paracrine signaling. However, our study highlights the fact that TNFα/TNFR1 is not the only death receptor ligand/receptor system that can mediate Smac mimetic-induced apoptosis, as the TNFα-blocking antibody Enbrel or TNFR1 knockdown largely fail to rescue BV6-induced apoptosis in A172 glioblastoma cells. It is noteworthy that we demonstrate that upregulation of DR5 can represent an additional mechanism besides TNFα/TNFR1 that is critical for Smac mimetic-triggered apoptosis. Independently of our study, there is very recent evidence of TNFα-independent, Smac mimetic-induced apoptosis in breast cancer cells, as neutralizing antibodies against TNFα or TNFR1 knockdown failed to reverse cell death.21 However, the molecular mechanisms of cell death induction by Smac mimetic were not identified in that study,21 underscoring the novelty of our present report.
Irrespectively of the differential requirement of TNFα/TNFR1 or DR5 signaling, our findings demonstrate that BV6-stimulated NF-κB activation is critically required for apoptosis induction, as inhibition of NF-κB blocks Smac mimetic-triggered apoptosis. These findings support a model according to which Smac mimetic, via depletion of IAP proteins, activates NF-κB that, in turn, engages NF-κB-dependent proapoptotic signaling pathways in a context-dependent manner via TNFα/TNFR1 or DR5 signaling. In addition, additional NF-κB-dependent, yet undefined, mechanisms may be involved.
Although Smac mimetic has been described to trigger NF-κB activation, the functional relevance of NF-κB as a pro- or antiapoptotic factor during Smac mimetic-induced apoptosis has been controversially discussed. On one side, activation of NF-κB was shown to be required for Smac mimetic-induced apoptosis, as inhibition of NF-κB abrogated Smac mimetic-induced apoptosis.22, 23, 24, 25 On the other side, an antiapoptotic function was ascribed to NF-κB, as blockage of NF-κB resulted in enhanced apoptosis upon treatment with Smac mimetic.26, 27 These differences may be explained by the context-dependent role of NF-κB in the regulation of apoptosis, as there is an increasing number of studies in recent years showing that NF-κB can exert a proapoptotic function under certain circumstances in a cell type- and/or stimulus-dependent manner in addition to its well-described antiapoptotic properties.28, 29, 30
Our present study provides further insights into the regulation of NF-κB signaling by Smac mimetic. Detailed analysis reveals activation of both canonical and non-canonical NF-κB pathways by BV6. This might be mediated through crosstalk between the two NF-κB signaling branches: NIK, a well-described mediator of non-canonical NF-κB signaling, accumulates upon cIAP1/2 depletion by BV6 and initiates p100 processing.12, 13 NIK is known to activate IKKα and can mediate crosstalk between the non-canonical and canonical branches of NF-κB signaling, for example, through activation of IKKβ, the main kinase for IκBα, resulting in IκBα degradation and activation of the canonical NF-κB pathway.7 Alternatively, Smac mimetic may directly activate canonical NF-κB signaling. Accordingly, the BV6-stimulated increase in the E3 ubiquitin ligase activity of cIAP proteins may trigger an initial wave of heteroubiquitination including ubiquitination of RIP1, leading to activation of the canonical NF-κB pathway. Our data support the first model, that is, activation of both NF-κB signaling branches via crosstalk, as hallmarks of canonical activation, such as IκBα phosphorylation, occur in parallel with the peak of NIK accumulation and well after degradation of cIAP1. Furthermore, our data demonstrate that dominant-negative IκBα-SR suppresses non-canonical NF-κB signaling via crosstalk mechanisms in addition to its well-known function as an inhibitor of the canonical NF-κB pathway. Accordingly, expression levels of p100 and RelB, the two NF-κB target genes, are suppressed upon IκBα-SR overexpression, thereby attenuating non-canonical NF-κB signaling.
By identifying DR5 as a critical mediator of Smac mimetic-induced apoptosis, our findings provide novel insights into the determinants that control susceptibility of cancer cells to Smac mimetic. The relevance of our study is underscored by the fact that small-molecule inhibitors of IAP proteins such as Smac mimetic are presently evaluated in early clinical trials.5 Thus, a better understanding of the molecular determinants of Smac mimetic-induced apoptosis will be critical for the successful translation of this strategy into clinical application.
Materials and Methods
Cell culture and chemicals
Cell lines were obtained from ATCC (Manassas, VA, USA) and grown in DMEM medium (Invitrogen, Karlsruhe, Germany) supplemented with 1% penicillin/streptomycin, 1% sodium pyruvate (both from Invitrogen) and 10% fetal calf serum (Invitrogen). BV6, the bivalent Smac mimetic,12 was kindly provided by Genentech (South San Francisco, CA, USA) and Enbrel by Pfizer (Berlin, Germany). Recombinant human TNFα was purchased from Biochrom (Berlin, Germany). All chemicals were obtained from Sigma (Deisenhofen, Germany) unless indicated otherwise.
Transduction and transfection
Overexpression of the dominant-negative IκBα-SR was performed by retroviral transduction using IκBα (S32; 36A) and the pCFG5-IEGZ retroviral vector system as previously described.29 Knockdown of DR5 was performed by lentiviral shRNA vectors as previously described.31 Shortly, HEK293T cells were transfected with 7.5 μg pGIPZ-shRNAmir vector, using calcium phosphate transfection. All pGIPZ-shRNAmir-vectors were purchased from Thermo Fisher Scientific (Dreieich, Germany): non-silencing control (Ctrl): RHS4346, shDR5_1 shRNA: RHS4430-99157936, shDR5_2 shRNA: RHS4430-101030035, shDR5_3 shRNA: RHS4430-101035311. Virus-containing supernatant was collected, filtered and used for spin transduction at 30°C in the presence of 8 μg/ml polybrene. Transduced cells were selected with 1 μg/ml puromycin (Sigma). For transient knockdown by siRNA, cells were reversely transfected with 5 nM SilencerSelect siRNA (Invitrogen), control siRNA (4390843) or targeting siRNAs (s16756 and s16758 for DR5, s14265 and s14266 for TNFR1 and s16663 for TRAIL) using Lipofectamine RNAi Max (Invitrogen) and OptiMEM (Life Technologies, Darmstadt, Germany).
Determination of apoptosis and cell viability
Apoptosis was determined by flow cytometric analysis of DNA fragmentation of propidium iodide (PI)-stained nuclei (FACSCanto II, BD Biosciences, Heidelberg, Germany) as described previously.32 The percentage of specific apoptosis was calculated by subtracting the percentage of spontaneous apoptosis from the percentage of induced apoptosis. Cell viability was assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay according to the manufacturer’s instructions (Roche Diagnostics, Mannheim, Germany).
Western blotting and immunoprecipitation
Western blot analysis was performed as described previously33 using the following antibodies: anti-cIAP1 (R&D Systems, Inc., Wiesbaden-Nordenstadt, Germany), anti-XIAP from BD Biosciences, anti-caspase-3, anti-NIK and anti-phospho-p65 from Cell Signaling (Beverly, MA, USA), anti-β-actin (Sigma), anti-α-tubulin (Calbiochem, Darmstadt, Germany), anti-p50, anti-p65, anti-RelB and anti-TNFR1 from Santa Cruz Biotechnology (Santa Cruz, CA, USA), anti-p52 and anti-DR5 (Millipore, Schwalbach, Germany), anti-lamin B1 (Abcam, Cambridge, UK) and anti-GAPDH (BioTrend, Cologne, Germany). Donkey anti-mouse IgG, donkey anti-rabbit IgG or donkey anti-goat IgG labeled with IRDye infrared dyes were used for fluorescence detection at 700 nm 800 nm (LI-COR Biotechnology, Bad Homburg, Germany). Caspases were immunodetected by enhanced chemoluminescence (Amersham Biosciences, Freiburg, Germany) using an anti-rabbit IgG-HRP as secondary antibody. Immunoprecipitation of caspase-8 and analysis of RIP1/FADD/caspase-8 interaction was performed as described previously.25
NF-κB reporter assay
NF-κB transcriptional activity was determined using the NF-κB Reporter System pTRH1-NF-κB-EGFP from System Biosciences (TR503PA-1, Mountain View, CA, USA) according to the manufacturer’s instructions. Briefly, NF-κB reporter activity was determined by flow cytometric analysis of median FITC intensity of the living cell population.
Quantitative real-time PCR
Total RNA was extracted using peqGOLD Total RNA kit from Peqlab Biotechnologie GmbH (Erlangen, Germany) according to the manufacturer’s instructions. Total RNA (2 μg) was used to synthetize the corresponding cDNA using RevertAid H Minus First Strand cDNA Synthesis Kit (MBI Fermentas GmbH, St. Leon-Rot, Germany). To quantify gene expression levels, SYBR-Green-based quantitative RT-qPCR was performed using the 7900HT fast real-time PCR system from Applied Biosystems (Darmstadt, Germany). Data were normalized on 28S-rRNA expression as reference gene. Primers are listed in Supplementary Table 1. Melting curves were plotted to verify the specificity of the amplified products. All determinations were performed in duplicate. The relative expression of the target gene transcript and reference gene transcript was calculated as ΔΔCt. At least two independent experiments were performed for each gene. TRAIL mRNA levels were assessed by TaqMan Gene Expression Assay (Life Technologies, Hs00921974_m1) according to the manufacturer’s protocol.
Statistical analysis
Statistical significance was assessed by two-sided Student’s t-test using Microsoft Excel (Microsoft Deutschland GmbH, Unterschleißheim, Germany); *P<0.05; **P<0.01.
Abbreviations
- BIR:
-
baculovirus IAP repeat
- cIAP1:
-
cellular inhibitor of apoptosis 1
- DISC:
-
death-inducing signaling complex
- DR5:
-
death receptor 5
- FADD:
-
FAS-associated protein with death domain
- GSEA:
-
gene set enrichment analysis
- IAP:
-
inhibitor of apoptosis
- IκBα-SR:
-
IκBα superrepressor
- MTT:
-
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NF-κB:
-
nuclear factor-κB
- PI:
-
propidium iodide
- RING:
-
really interesting new gene
- RIP1:
-
receptor-interacting protein 1
- RT-qPCR:
-
reverse transcriptase quantitative PCR
- Smac:
-
second mitochondria-derived activator of caspase
- TNF:
-
tumor necrosis factor
- TNFR1:
-
tumor necrosis factor receptor 1
- TRAF:
-
TNF receptor-associated factor
- TRAIL:
-
TNF-related apoptosis-inducing ligand
- XIAP:
-
X-linked inhibitor of apoptosis protein
- zVAD.fmk:
-
Z-Val-Ala-Asp (OMe)-fluoromethylketone
References
Lockshin RA, Zakeri Z . Cell death in health and disease. J Cell Mol Med 2007; 11: 1214–1224.
Fulda S . Tumor resistance to apoptosis. Int J Cancer 2009; 124: 511–515.
Ashkenazi A . Targeting the extrinsic apoptosis pathway in cancer. Cytokine Growth Factor Rev 2008; 19: 325–331.
Chu WM . Tumor necrosis factor. Cancer Lett 2013; 328: 222–225.
Fulda S, Vucic D . Targeting IAP proteins for therapeutic intervention in cancer. Nat Rev Drug Discov 2012; 11: 109–124.
Eckelman BP, Salvesen GS, Scott FL . Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep 2006; 7: 988–994.
Oeckinghaus A, Hayden MS, Ghosh S . Crosstalk in NF-kappaB signaling pathways. Nat Immunol 2011; 12: 695–708.
Varfolomeev E, Goncharov T, Fedorova AV, Dynek JN, Zobel K, Deshayes K et al. c-IAP1 and c-IAP2 are critical mediators of tumor necrosis factor alpha (TNFalpha)-induced NF-kappaB activation. J Biol Chem 2008; 283: 24295–24299.
Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell 2008; 30: 689–700.
Vallabhapurapu S, Matsuzawa A, Zhang W, Tseng PH, Keats JJ, Wang H et al. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat Immunol 2008; 9: 1364–1370.
Zarnegar BJ, Wang Y, Mahoney DJ, Dempsey PW, Cheung HH, He J et al. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat Immunol 2008; 9: 1371–1378.
Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell 2007; 131: 669–681.
Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell 2007; 131: 682–693.
Wang L, Du F, Wang X . TNF-alpha induces two distinct caspase-8 activation pathways. Cell 2008; 133: 693–703.
Bai L, McEachern D, Yang CY, Lu J, Sun H, Wang S . LRIG1 modulates cancer cell sensitivity to Smac mimetics by regulating TNFalpha expression and receptor tyrosine kinase signaling. Cancer Res 2012; 72: 1229–1238.
Ravi R, Bedi GC, Engstrom LW, Zeng Q, Mookerjee B, Gelinas C et al. Regulation of death receptor expression and TRAIL/Apo2L-induced apoptosis by NF-kappaB. Nat Cell Biol 2001; 3: 409–416.
Shetty S, Graham BA, Brown JG, Hu X, Vegh-Yarema N, Harding G et al. Transcription factor NF-kappaB differentially regulates death receptor 5 expression involving histone deacetylase 1. Mol Cell Biol 2005; 25: 5404–5416.
Chen JJ, Chou CW, Chang YF, Chen CC . Proteasome inhibitors enhance TRAIL-induced apoptosis through the intronic regulation of DR5: involvement of NF-kappa B and reactive oxygen species-mediated p53 activation. J Immunol 2008; 180: 8030–8039.
Petersen SL, Wang L, Yalcin-Chin A, Li L, Peyton M, Minna J et al. Autocrine TNFalpha signaling renders human cancer cells susceptible to Smac-mimetic-induced apoptosis. Cancer Cell 2007; 12: 445–456.
Elias A, Siegelin MD, Steinmuller A, von Deimling A, Lass U, Korn B et al. Epigenetic silencing of death receptor 4 mediates tumor necrosis factor-related apoptosis-inducing ligand resistance in gliomas. Clin Cancer Res 2009; 15: 5457–5465.
Allensworth JL, Sauer SJ, Lyerly HK, Morse MA, Devi GR . Smac mimetic Birinapant induces apoptosis and enhances TRAIL potency in inflammatory breast cancer cells in an IAP-dependent and TNF-alpha-independent mechanism. Breast Cancer Res Treat 2013; 137: 359–371.
Wagner L, Marschall V, Karl S, Cristofanon S, Zobel K, Deshayes K et al. Smac mimetic sensitizes glioblastoma cells to Temozolomide-induced apoptosis in a RIP1- and NF-kappaB-dependent manner. Oncogene 2013; 32: 988–997.
Stadel D, Cristofanon S, Abhari BA, Deshayes K, Zobel K, Vucic D et al. Requirement of nuclear factor kappaB for Smac mimetic-mediated sensitization of pancreatic carcinoma cells for gemcitabine-induced apoptosis. Neoplasia 2011; 13: 1162–1170.
Berger R, Jennewein C, Marschall V, Karl S, Cristofanon S, Wagner L et al. NF-{kappa}B is required for Smac mimetic-mediated sensitization of glioblastoma cells for {gamma}-irradiation-induced apoptosis. Mol Cancer Ther 2011; 10: 1867–1875.
Abhari BA, Cristofanon S, Kappler R, von Schweinitz D, Humphreys R, Fulda S . RIP1 is required for IAP inhibitor-mediated sensitization for TRAIL-induced apoptosis via a RIP1/FADD/caspase-8 cell death complex. Oncogene 2013; 32: 3263–3273.
Dai Y, Liu M, Tang W, Li Y, Lian J, Lawrence TS et al. A Smac-mimetic sensitizes prostate cancer cells to TRAIL-induced apoptosis via modulating both IAPs and NF-kappaB. BMC Cancer 2009; 9: 392.
Bai L, Chen W, Wang X, Tang H, Lin Y . IKKbeta-mediated nuclear factor-kappaB activation attenuates smac mimetic-induced apoptosis in cancer cells. Mol Cancer Ther 2009; 8: 1636–1645.
Perkins ND . The diverse and complex roles of NF-kappaB subunits in cancer. Nat Rev Cancer 2012; 12: 121–132.
Karl S, Pritschow Y, Volcic M, Hacker S, Baumann B, Wiesmuller L et al. Identification of a novel pro-apopotic function of NF-kappaB in the DNA damage response. J Cell Mol Med 2009; 13: 4239–4256.
Jennewein C, Karl S, Baumann B, Micheau O, Debatin KM, Fulda S . Identification of a novel pro-apoptotic role of NF-kappaB in the regulation of TRAIL- and CD95-mediated apoptosis of glioblastoma cells. Oncogene 2012; 31: 1468–1474.
Gonzalez P, Mader I, Tchoghandjian A, Enzenmuller S, Cristofanon S, Basit F et al. Impairment of lysosomal integrity by B10, a glycosylated derivative of betulinic acid, leads to lysosomal cell death and converts autophagy into a detrimental process. Cell Death Differ 2012; 19: 1337–1346.
Fulda S, Friesen C, Los M, Scaffidi C, Mier W, Benedict M et al. Betulinic acid triggers CD95 (APO-1/Fas)- and p53-independent apoptosis via activation of caspases in neuroectodermal tumors. Cancer Res 1997; 57: 4956–4964.
Vogler M, Durr K, Jovanovic M, Debatin KM, Fulda S . Regulation of TRAIL-induced apoptosis by XIAP in pancreatic carcinoma cells. Oncogene 2007; 26: 248–257.
Acknowledgements
We thank D Vucic (Genentech, South San Francisco, CA, USA) for providing BV6, J Silke (Melbourne, Australia) for providing the NF-κB reporter plasmid, K Strebhardt for providing MDA-MB-231 cells, R Sauter for expert technical assistance and C Hugenberg for expert secretarial assistance. This work has been partially supported by grants from the Deutsche Forschungsgemeinschaft and IAPVII (to S F).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Raschellá
Supplementary Information accompanies this paper on Cell Death and Disease website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Eckhardt, I., Roesler, S. & Fulda, S. Identification of DR5 as a critical, NF-κB-regulated mediator of Smac-induced apoptosis. Cell Death Dis 4, e936 (2013). https://doi.org/10.1038/cddis.2013.457
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.457
Keywords
This article is cited by
-
BH3-mimetics: recent developments in cancer therapy
Journal of Experimental & Clinical Cancer Research (2021)
-
Smac mimetic suppresses tunicamycin-induced apoptosis via resolution of ER stress
Cell Death & Disease (2019)
-
Elucidation for modulation of death receptor (DR) 5 to strengthen apoptotic signals in cancer cells
Archives of Pharmacal Research (2019)
-
Copper-imidazo[1,2-a]pyridines induce intrinsic apoptosis and modulate the expression of mutated p53, haem-oxygenase-1 and apoptotic inhibitory proteins in HT-29 colorectal cancer cells
Apoptosis (2019)
-
NF-κB contributes to Smac mimetic-conferred protection from tunicamycin-induced apoptosis
Apoptosis (2019)