Abstract
Autophagy is a critical regulator of organellar homeostasis, particularly of mitochondria. Upon the loss of membrane potential, dysfunctional mitochondria are selectively removed by autophagy through recruitment of the E3 ligase Parkin by the PTEN-induced kinase 1 (PINK1) and subsequent ubiquitination of mitochondrial membrane proteins. Mammalian sequestrome-1 (p62/SQSTM1) is an autophagy adaptor, which has been proposed to shuttle ubiquitinated cargo for autophagic degradation downstream of Parkin. Here, we show that loss of ref(2)P, the Drosophila orthologue of mammalian P62, results in abnormalities, including mitochondrial defects and an accumulation of mitochondrial DNA with heteroplasmic mutations, correlated with locomotor defects. Furthermore, we show that expression of Ref(2)P is able to ameliorate the defects caused by loss of Pink1 and that this depends on the presence of functional Parkin. Finally, we show that both the PB1 and UBA domains of Ref(2)P are crucial for mitochondrial clustering. We conclude that Ref(2)P is a crucial downstream effector of a pathway involving Pink1 and Parkin and is responsible for the maintenance of a viable pool of cellular mitochondria by promoting their aggregation and autophagic clearance.
Similar content being viewed by others
Main
Accumulating evidence suggests that the disruption of mitochondrial function and dynamics contributes to age-related neurodegenerative diseases such as Parkinson’s disease (PD).1 Accordingly, cells have developed complex quality-control mechanisms to cope with the different challenges constantly imposed on the integrity of mitochondria.
Recent findings have demonstrated that proteins coded by two genes mutated in familial PD, PTEN-induced kinase 1 (PINK1) and the E3 ubiquitin ligase PARKIN are components of a pathway that mediates the autophagic disposal of defective mitochondria.2 In damaged mitochondria, PINK1 recruits PARKIN to catalyse the ubiquitination of proteins on the outer mitochondrial membrane, such as the voltage-dependent anion channel 1 (VDAC1)3 and Mitofusin (Mfn).4
Mammalian p62 is an autophagy adaptor that can interact with ubiquitin conjugated to a target protein as well as LC3/GABARAP on autophagosomes.5 Following mitochondrial damage, p62 can recognise Parkin-mediated, poly-ubiquitinated chains in mitochondrial clusters and mediate their autophagic clearance.3 Refractory to Sigma P, ref(2)P, the single Drosophila orthologue of P62, has been recently shown to have a key role in parkin-mediated suppression of mitochondrial dysfunction, which is induced by protein conformational stress,6 suggesting that Ref(2)P might have an active role in mitochondrial quality control. Here, we investigate the consequences of ref(2)P mutations on mitochondrial integrity and genetically address its role in mitochondrial quality control through the Pink1/Parkin pathway.
Results
ref(2)P mutants exhibit mitochondrial defects
The Drosophila orthologue of P62, ref(2)P, has been shown to act in the formation of ubiquitin-positive protein aggregates under physiological conditions and when normal protein turnover is inhibited.7 Ref(2)P contains three domains involved in protein–protein interactions, a PB1 domain involved in multimerisation, a ZZ zinc-finger and an ubiquitin-associated domain (UBA). Drosophila ref(2)Pod2 and ref(2)Pod3mutants lack the PB1 and UBA domains, respectively (Figure 1a), and both are viable and reported to be completely male-sterile.8 As mitochondrial function is required for normal spermatogenesis and dysfunctional mitochondria are the basis of infertility in pink1 and parkin null flies, we examined morphological changes in mitochondria during spermatogenesis in ref(2)Pod2 and ref(2)Pod3mutants. In both mutants, cysts of the early onion-stage spermatids presented with uniformly sized nuclei, associated with abnormal mitochondrial derivatives (also known as nebenkern) (Figures 1b–e). In the spermatids of ref(2)Pod2 mutants, the nebenkern enveloped the white nuclei instead of being adjacent to them (Figure 1c), as is the case in the control spermatids (Figure 1b). Despite the conserved position of the nebenkern in the ref(2)Pod3spermatids, they appeared pale when compared with the densely packed spherical shapes present in the control spermatids (Figure 1d).
Characterisation of ref(2)P-mutant flies. (a) Structural domain organisation of mammalian p62, Drosophila Ref(2)P, and Ref(2)Pod2 and Ref(2)Pod3 mutants. p62 has a PB1 domain, a ZZ-type zinc-finger domain, a TRAF6-binding (TB) domain, an LC3-interacting region (LIR) and a UBA. Ref(2)P has in common with the mammalian p62, the PB1 domain, the ZZ-type finger domain and the UBA domain. Ref(2)Pod2 mutant protein lacks the PB1 domain, and Ref(2)Pod3 mutant protein lacks the UBA domain. (b–e) Schematic and phase contrast micrographs of mitochondrial morphogenesis in spermatids during the ‘onion stage’. Mutations in ref(2)P lead to morphological defects in the mitochondrial derivative (Nebenkern) of the onion-stage early spermatids. ref(2)Pod2 mutants present an atypical localisation of the nebenkern, surrounding the white nuclei (c). ref(2)Pod3 mutants exhibit an abnormally pale mitochondrial derivative (d). Scale bar, 50 μm (f–k) TEM images of single post-individualisation cysts containing 64 spermatids. (f and i) In the control (w1118), each spermatid contains an axoneme (arrowhead) and mitochondrial derivative (arrow) within an individual plasma membrane. Both ref(2)P mutant cyst show individualisation defects. (g and j) In ref(2)Pod2 mutants, the mitochondrial derivative appears bigger and contains a large mass of electron-dense material (arrow); (h and k) the ref(2)Pod3 mutant cysts have an overall disorganised architecture, with a rather normal axoneme (arrowhead) but a large vacuolised mitochondrial derivative (arrow). Scale bar, 2 μm (f–h), 500 nm (i–k)
To further characterise these mitochondrial defects, we examined the ref(2)P mutant testes using transmission electron microscopy (TEM). Following the onion stage, Drosophila spermatids proceed to the elongation and maturation stages. After elongation, the spermatids undergo a process known as individualisation, during which the connections between the 64 spermatids in a cyst are lost and most of the spermatid cytoplasm is expelled. After individualisation is completed, each spermatid largely consists of the central axoneme, a microtubule-based cytoskeleton that is essential for motility (arrowheads, Figures 1f and i), and the mitochondrial derivatives (arrows, Figures 1f and i), which are adjacent to the axoneme. Curiously, we found significant differences between the two ref(2)P mutants in this individualisation process. Spermatids in the ref(2)Pod2 mutant testes underwent normal elongation; however, the individualisation process was defective. Although the numbers of spermatids in each cyst, as well as the shape and size of the axoneme, were comparable to those in the controls, the spatial relationship between the axoneme and mitochondria was affected. There was an increased distance between the two structures and the mitochondria seemed to contain a large mass of electron-dense material, which may represent aberrant mitochondria (arrows, Figures 1g and j). As for the ref(2)Pod3 mutant, the overall architecture of the cysts was completely disorganised (Figures 1h). Here, the axonemes were easily visible (arrowheads, Figures 1h and k); however, the mitochondria were frequently smaller than those in the control cysts, containing electron-dense material and a striking vacuolisation (arrows, Figures 1h and k).
The striking morphological defects observed in the sperm cell mitochondria of ref(2)P mutant flies, together with the proposed role for Ref(2)P in the clearance of damaged mitochondria,6 led us to test whether its loss might be associated with defects in the removal of defective mitochondria. The qualitative analysis of the spatial distribution of mitochondrial DNA (mtDNA) nucleoids in the indirect flight muscle of ref(2)P mutants revealed an increase in their density (Figure 2a). We next performed a quantitative analysis of the levels of mtDNA in both control and ref(2)p mutants and found these to be increased in both young and aged ref(2)Pod2 and ref(2)Pod3 mutants (Figure 2b). To determine whether the enhanced mtDNA levels in ref(2)P mutants was linked to a higher mitochondrial density (MD), we assessed this parameter in both control and ref(2)P mutants. This was performed by analysing the activity of the mitochondrial matrix enzyme citrate synthase (CS), an indirect measurement of MD,9 as well as by measuring the levels of Cytochrome c (Cyt c) and mitochondrial transcription factor 1 (mtTFA). We failed to detect any significant changes in either the activity of CS (Figure 2c), the levels of Cyt c (Figure 2d) or the levels of mtTFA (Figure 2e), indicating that ref(2)P mutants do not have an increase in MD. We next assessed whether the loss of ref(2)P leads to mitochondrial impairment. To achieve this, we compared the respiration rates in controls and ref(2)P mutants. High-resolution analysis using whole flies failed to detect any significant changes in the respiration rates of ref(2)Pod2 and ref(2)Pod3 mutants (Figure 2f).
Mutations in ref(2)P result in the accumulation of mtDNA. (a) Regular mitochondrial distribution is disrupted in ref(2)P mutants and the number of mtDNA nucleoids (mt) is increased. PicoGreen also labels the dsDNA of muscle nuclei (nuc). (b) ref(2)P mutants flies showed an increase in mtDNA. The ratio of mtDNA to nDNA was measured using real-time quantitative PCR in flies with the indicated ages (mean±S.D., n=9 per genotype). Statistically significant values relative to the control (w1118) are indicated by asterisks (one-way ANOVA with Dunnett’s multiple comparison test). (c and d) Mutations in ref(2)P do not cause alterations in mitochondrial protein density. Protein density was determined in 26-day-old flies with the indicated genotypes by determining the concentration of Cyt c using an ELISA assay (d) and by measuring the activity of the mitochondrial matrix enzyme citrate synthase (c). Data are shown as the mean ±S.E.M (n=3 in each group). Statistical significance relative to the control (w1118) is indicated (one-way ANOVA with Dunnett’s multiple comparison test). (e) Analysis of the mtTFA levels in ref(2)P mutants. Lysates prepared from adult flies were subjected to western blot analysis with the indicated antibodies. (f) Normal respiration in ref(2)P mutant flies. Activity of the indicated complexes in coupled mitochondria was measured in 3-day-old flies using high-resolution respirometry. Data are shown as the mean±S.E.M. (n≥4 in each genotype). Statistical significance relative to the control (w1118) is indicated (one-way ANOVA with Dunnett’s multiple comparison test)
ref(2)P mutants exhibit increased levels of heteroplasmic mtDNA mutations
Defects in mitophagy, the autophagic disposal of defective mitochondria, lead to the accumulation of damaged organelles and result in the enhanced production of DNA-damaging agents such as reactive oxygen species (ROS) that can cause mtDNA mutations. As the mitochondrial genome is present in multiple copies in each organelle,10 mutant mitochondrial genomes can co-exist with normal genomes in a situation known as heteroplasmy. This implies that once a critical threshold of mutant DNA copies is reached, normal mitochondrial function may be altered. To address whether the increased levels of mtDNA observed in ref(2)P mutants were associated with enhanced heteroplasmy in mtDNA because of possible defects in mitophagy we used Next-Generation Sequencing (NGS) technology to sequence the mitochondrial genome in control and ref(2)P mutant flies. A significant percentage of the mitochondrial genome was analysed at high coverage, with each of the sequenced mtDNA bases being sequenced, on average, 4 400 times. This permitted us to identify heteroplasmic variants in specific regions of the mtDNA genome (Figure 3a). The outcome of this analysis revealed an increase in heteroplasmy in both young (3 days old) and aged (25 days old) ref(2)P mutants (Figure 3b), suggesting that defects in autophagy in ref(2)P mutants are associated with increased heteroplasmy in the mitochondrial genome of ref(2)P mutants.
Detection of heteroplasmic mtDNA variants in Drosophila ref(2)P mutants. (a) Next-Generation Sequencing analysis of the Drosophila mitochondrial genome. Horizontal scale corresponds to the position of individual features in the Drosophila mitochondria genome. Vertical scale indicates the average read depth obtained for each individual base. Genes and features shown in red indicate areas where heteroplasmy was detected in individual genomes. The circle indicates the origin of replication for the mitochondrial genome (mt:ori) and is located to the left of the A+T-rich region of the mitochondrial genome. (b) Heteroplasmic variants in the mtDNA of adult flies. Frequencies of 1.6% or more of the variant allele when compared with the reference allele30 are shown
ref(2)P mutants show decreased lifespan and an age-dependent decrease in motor performance
To further evaluate the phenotypic consequences of mitochondrial abnormalities in ref(2)P mutants, we next analysed their overall survival. We observed a decrease in the lifespan of ref(2)Pod2 and ref(2)Pod3 mutants maintained on normal food (Figure 4a) as well as in food containing rotenone, a mitochondrial complex I inhibitor (Figure 4b) or paraquat, a pesticide linked to PD by epidemiological studies11 (Figure 4c). Mitochondrial impairment such as that observed in pink1-mutant flies has been shown to cause locomotor defects by inducing pathologies in the Drosophila skeletal muscles, including the indirect flight muscles.12, 13 We have previously shown that ref(2)P mutants are also characterised by mitochondrial defects in the indirect flight muscles.6
ref(2)P mutant flies have a decreased lifespan and motor impairment. (a–c) Fly viability was scored over the indicated periods, using a minimum of 80 flies per genotype. Statistical significance is indicated (log-rank, Mantel–Cox test). (d) Mutations in ref(2)P result in significant motor impairment. Flies with the indicated genotypes and ages were tested using a standard climbing assay (mean ±S.D., n≥100 flies for each genotype)). Statistical significance relative to the control (w1118) is indicated (two-way ANOVA with Dunnett’s multiple comparison test)
Both ref(2)P mutants showed a strong impairment in the locomotor activity from a very young age that progressed with age (Figure 4d).
Expression of ref(2)P suppresses mitochondrial dysfunction in the pink1 mutants
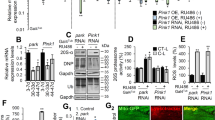
The defective mitochondrial phenotypes observed in the ref(2)P mutants were reminiscent of pink1 mutant flies; these phenotypes included abnormal mitochondrial morphology in the indirect flight muscle,6 reduced climbing ability, reduced lifespan and mitochondria-linked abnormalities of the sperm cells. We therefore decided to test the epistatic relationship between ref(2)P and pink1. Using the UAS-GAL4 system,14 we generated transgenic flies expressing the full-length ref(2)P, and after confirming the transgene expression using western blot analysis (Figure 5a), we determined that ref(2)P expression reverts the male sterility observed in ref(2)Pod3mutants (Figure 5b). Next, we determined the effect of the enhanced ubiquitous ref(2)P expression in pink1 mutants. We observed that the thoracic indentation phenotype exhibited in the pink1B9 mutants13 (Figure 5c) was markedly suppressed by ref(2)P expression (Figure 5d). Ultrastructural analysis of the indirect flight muscles revealed a similar recovery of tissue structure and mitochondrial integrity (Figures 5e–g). This recovery was also reflected in an improved climbing ability (Figure 5h) and a rescue of mitochondrial protein content, assessed through analysis of the levels of Complex I and V (Figure 5i). The main features of pink1 mutant flies are the degeneration of both the indirect flight muscle and dopaminergic neurons.13 The loss of synaptic mitochondria has been linked to defects in neurotransmission in the Drosophila neuromuscular junction (NMJ).15 To test whether the degeneration of the indirect flight muscle in pink1 mutant flies involves a presynaptic component, we investigated the effects of the neuronal expression of ref(2)P. pink1B9 mutants exhibit age-related degeneration of a subset of dopaminergic neurons,13 including the protocerebral posterior lateral 1 (PPL1) cluster.16 The ectopic expression of ref(2)P using the pan-neural elav-GAL4 driver rescued the dopaminergic neuron loss in the PPL1 cluster of pink1 mutants (Figure 5j). Notably, the neuronal expression of ref(2)P in pink1 mutants significantly decreased the presence of the crushed thorax phenotype of pink1 mutants (Figure 5k). These effects indicate that the neuronal rescue of mitochondrial function in pink1 mutants is sufficient to suppress indirect flight muscle degeneration and that the muscle defects in these mutants have a presynaptic component.
ref(2)P gain-of-function rescues pink1 mutant flies. (a) Analysis of Ref(2)P levels. Whole-fly lysates were analysed using western blotting with the indicated antibodies. (b) Reduced fertility in ref(2)Pod3 mutants is rescued by expression of ref(2)P. Single male of the indicated genotypes were mated with w1118 virgin females for 24 h before the males were removed. Total number of vials with progenies per genotype were scored (n). (c) Thoracic defects of pink1 mutants. SEM micrographs of thoraces from flies with the indicated genotypes show that the collapsed-thorax phenotype of pink1 mutants. A control thorax (w1118) is also shown. (d) Expression of ref(2)P rescued the thoracic indentations of pink1 mutants (n≥60 flies per genotype). Asterisk indicates statistical significance relative to pink1B9, da>Ref(2)P (χ2). (e–g) ref(2)P expression rescues the mitochondrial abnormalities observed in the indirect flight muscles of the pink1 null flies. Tissues were analysed using TEM. Scale bar, 2 μm. (h) Expression of ref(2)P rescued the climbing defects of pink1 flies. Flies were tested using a standard climbing assay (mean ±S.D., n≥60 flies per genotype). Asterisks indicate statistically significant values relative to the control (daGal4/+) (one-way ANOVA with Dunnett’s multiple comparison test). (i) Expression of ref(2)P restored mitochondrial Complex I and V protein levels in pink1 mutant flies. Whole-fly protein lysates were subjected to western blot analysis with the indicated antibodies. (j) Neuronal expression of ref(2)P rescued the loss of dopaminergic neurons in the PPL1 cluster of pink1 mutant flies (mean ±S.D., n≥16 flies per genotype). The asterisks indicate statistically significant values (one-way ANOVA with Bonferroni’s multiple comparison test) relative to pink1B9. (k) Neuronal expression of ref(2)P rescued the thoracic defects of pink1 mutants (n≥76 flies per genotype). Asterisk(s) indicate statistical significance (Fisher’s exact test)
ref(2)P is required for mitochondrial clustering and parkin-mediated suppression of the pink1 mutant phenotype
We have previously shown that Ref(2)P is required for the Parkin-mediated suppression of defects associated with mitochondrial protein misfolding,6 suggesting its importance in clearing damaged mitochondria.
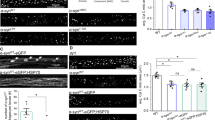
Mitochondrial dysfunction in pink1 mutants is associated with clustering of these organelles in the indirect flight muscle (Figure 6b). Analysis of the pink1 and ref(2)P double mutants showed a significant suppression of mitochondrial clustering in the indirect flight muscle in the pink1B9, ref(2)Pod2mutants (Figure 6c) and, to a greater extent, in the pink1B9, ref(2)Pod3 mutants (Figure 6d), indicating that Ref(2)P is required for the aggregation of defective mitochondria.
Drosophila ref(2)P is required for mitochondrial aggregation and parkin-mediated suppression of the pink1 mutant phenotype. (a–d) The Ref(2)P PB1 and UBA domains are essential for mitochondrial aggregation in Drosophila. Mitochondria were visualised by confocal analysis of the indirect flight muscles in a mitochondria-targeted protein (mitoGFP) transgenic animals with the da-GAL4 driver. Arrows point to aggregated mitochondria. Purple: nuclei, green: mitoGFP. Scale bar, 20 μm. (e) ref(2)P mutations blocked the Parkin-mediated suppression of the thoracic indentations in pink1 flies. Thoracic indentations were scored in flies of each indicated genotype (n≥60 flies per genotype). Asterisks indicate the statistical significance relative to pink1B9, da>Parkin (χ2). (f) ref(2)P mutations blocked the Parkin-mediated suppression of mitochondrial loss in pink1 mutant flies. Whole-fly lysates were analysed using western blotting with the indicated antibodies
Both parkin and pink1 are known to genetically interact in Drosophila, and Parkin and p62 were shown to physically interact in mammalian cells.6 We therefore decided to test the role of Ref(2)P in the Parkin-mediated suppression of the pink1 mutant phenotype. Parkin expression markedly reduced both the degree of thoracic indentations and the loss of mitochondrial proteins in pink1 mutants. However, this effect was reduced in the pink1B9, ref(2)Pod2 mutants and, to a greater extent, in the pink1B9, ref(2)Pod3 mutants (Figures 6e and f). As with the mitochondrial aggregation, the failure of parkin to suppress the pink1 phenotype was more profound in the ref(2)Pod3 than in the ref(2)Pod2 mutant background.
ref(2)P and parkin-mediated autophagy rescue pink1 mutant flies
Both PINK1 and Parkin are components of a stress-response pathway that targets defective mitochondria for selective autophagic degradation.17
ATG1 is a crucial regulator of autophagy both in yeast and Drosophila.18, 19, 20, 21 To test the requirement for autophagy in Parkin-mediated suppression of the phenotype of pink1 mutant flies, we utilised an atg1 mutant strain. In the atg1 and pink1 double-mutant flies, parkin failed to rescue the thoracic indentations (Figure 7a), the mitochondrial protein loss (Figure 7b) or the climbing defects (Figure 7c) observed in the pink1 mutant flies. These observations indicate that Parkin-mediated rescue of the mitochondrial defects caused by the loss of Pink1 is autophagy-dependent.
Autophagy is required for parkin- and ref(2)P-mediated suppression of the pink1 mitochondrial phenotype. Autophagy is required for parkin-mediated (a) suppression of thoracic indentations (n≥60 flies per genotype), asterisk indicates statistical significance relative to pink1B9, da>Parkin (χ2); (b) recovery of mitochondrial loss, flies were analysed using western blotting using the indicated antibodies; and (c) rescuing of the motor impairment in pink1 mutants, flies with the indicated genotypes were tested using a standard climbing assay (mean ±S.D., n≥60 flies for each genotype), asterisks indicate statistically significant values relative to the control (daGal4/+) (one-way ANOVA with Dunnett’s multiple comparison test). Autophagy is also essential for ref(2)P-mediated (d) suppression of thoracic indentations in pink1 flies, asterisk indicates statistical significance relative to pink1B9, da>Ref(2)P (χ2); and (e) climbing defects observed in pink1 mutants. Flies were tested using a standard climbing assay (mean±S.D., n≥60 flies per genotype); asterisks indicate statistically significant values relative to pink1B9 (one-way ANOVA with Dunnett’s multiple comparison test). Contrary to the pink1 mutants, ref(2)P failed to suppress the thoracic indentations in parkin flies (d, green)
As our genetic data show that ref(2)P interacts with Pink1 and acts as a downstream mediator of parkin function, we next tested whether autophagy was essential for the protective effects of ref(2)P. In the atg1 mutant background, ref(2)P was no longer able to suppress the thoracic indentations (Figure 7d) or the climbing defects (Figure 7e) observed in pink1 mutants. Taken together, these findings indicate that autophagy is a critical downstream effector of the protective effect of Parkin and Ref(2)P, which both act in autophagic clearance of mitochondria in flies.
Discussion
Mitochondrial dysfunction has been strongly associated with different neurodegenerative diseases, such as PD. Cells within complex multicellular organisms have developed quality-control mechanisms to cope with the many challenges that are constantly imposed on mitochondria and to suppress the accumulation of dysfunctional organelles.22
Here, we provide evidence that the Drosophila orthologue of p62, Ref(2)P is an important component of the Pink1/Parkin quality-control pathway.
ref(2)P mutant flies exhibited several pathological and functional phenotypes reminiscent of those observed in the pink1 or parkin mutants. They exhibited the following: mitochondrial abnormalities of the sperm cells; defective locomotor activity; and a shorter lifespan. These defective phenotypes are more profound in the ref(2)P mutants lacking the UBA domain, suggesting that the ability of Ref(2)P to bind ubiquitinated targets is required for mitochondrial integrity and function. Despite the observed mitochondrial defects in the ref(2)Pod2 and ref(2)Pod3 mutants, we did not detect any global alterations in mitochondrial mass or function, suggesting that mitochondrial dysfunction in the ref(2)P mutants is not as pronounced as that observed in pink1 or parkin mutant flies. The observed decrease in motor performance from an early age suggests that any mitochondrial defects in ref(2)P mutants might preferentially affect tissues with high energetic demand such as the skeletal muscles and spermatids. It is therefore possible that such defects are undetectable in the respirometry assays, as these are performed using mitochondria derived from whole flies. There have been a number of reports of mtDNA point mutations associated with neurodegenerative diseases such as PD.23 Here, we show for the first time that defects in ref(2)P, the single Drosophila P62 orthologue, result in an increase in mtDNA heteroplasmy. It is therefore conceivable that defects in mitophagy might contribute to neurodegenerative diseases such as PD by increasing the load of deleterious mtDNA mutations, leading eventually to increased mitochondrial dysfunction and an impairment of neuronal function.
ref(2)P mutants exhibited a marked sensitivity to rotenone, an organic pesticide that directly targets respiration by inhibiting mitochondrial complex I. These mutants also showed a lower sensitivity to paraquat, an herbicide widely used in agriculture that has been linked to PD. Paraquat increases oxidative stress, whereas rotenone causes mitochondrial dysfunction; however, both processes are linked and both pesticides affect these mechanisms (reviewed in Kamel24). Paraquat does not directly target mitochondria. In cells, it undergoes redox cycling in vivo to produce superoxide-free radicals that can damage not only these organelles but also other cellular components. We therefore reason that, within this context, the Ref(2)P-dependent mitophagy might be particularly important in suppressing damage caused by PD-linked toxins whose mechanism of action directly targets mitochondrial function such as rotenone.
We noted that expression of ref(2)P is capable of suppressing pink1 but not parkin (Figure 7d) mutant phenotypes. This finding indicates that Ref(2)P exerts a protective effect downstream of Pink1 but requires the presence of functional Parkin. This is compatible with a model in which Ref(2)P recognises mitochondrial substrates ubiquitinated by Parkin and therefore, in the absence of Parkin, is incapable of recognising ubiquitin-decorated mitochondria and targeting them for autophagy. Parkin failed to restore the mitochondrial function of pink1 mutant flies in the absence of functional Ref(2)P, supporting the notion that Ref(2)P is a critical downstream effector of Parkin.
Mutations in ref(2)P suppressed mitochondrial aggregation in the pink1 mutant flies. In mammalian cells, p62 has been suggested to mediate the aggregation of dysfunctional mitochondria into tight clusters, thereby minimising the surface area exposed to other cellular components.25, 26 Here, we show that this function of p62 is conserved in Drosophila and that Ref(2)P coordinates mitochondrial clustering through its PB1 and UBA domains. Reducing the surface area of impaired mitochondria within the cell by mitochondrial clustering may help minimise the uptake of respiratory substrates and limit the spread of mitochondrial ROS to other cellular compartments. Additionally, the clustering of the dysfunctional mitochondria could be beneficial to subcellular compartments with high-energy requirements, such as neuronal synapses, by preventing damaged mitochondria from being transported at the expense of bioenergetically active mitochondria. Alternatively, as p62 clustering of ubiquitinated substrates has been shown to cause cell death in the absence of its autophagic degradation,27 it is possible that, in Drosophila, Ref(2)P functions as a sensor of defective mitophagy quality control, triggering cell death when the removal of Parkin-ubiquitinated mitochondria is insufficient. This scenario could explain the suppression of the pink1 mutant phenotypes by ref(2)P expression.
Finally, both Parkin and p62 are important regulators of mitophagy. Parkin is responsible for the autophagic elimination of damaged mitochondrial units.2 p62, on the other hand, binds directly to the autophagy protein LC3 and is believed to serve as an autophagy receptor for ubiquitinated protein aggregates as well as peroxisomes and intracellular bacteria.5 Our data provide robust genetic evidence that inhibiting autophagy through mutations in Drosophila atg1 prevents both Parkin and Ref(2)P from exerting their protective effects on mitochondria.
Our data indicate that enhancing the autophagy pathway improved mitochondrial function in a Drosophila model of mitochondrial dysfunction, suggesting this pathway and clearance of damaged mitochondria as a potential therapeutic target in PD pathogenesis. This opens a promising avenue of exploring the role of autophagy-inducing agents in the prevention and treatment of neurodegenerative diseases, such as PD associated with mitochondrial dysfunction.
Materials and Methods
Antibodies
The primary antibodies used in this study were anti-Complex I subunit NDUFS3 (Mitosciences, Eugene, OR, USA, Cat. No. MS112), mtTFA (Abcam, Cambridge, UK, Cat. No. ab47548), anti-α-Tubulin (Sigma-Aldrich, Cat. No. T6054), anti-ATP synthase subunit alpha (Mitosciences, Cat. No. MS507) and anti-HA (Roche Applied Science, Indianapolis, IN, USA, Cat. No. 11583816001).
Genetics and Drosophila strains
Transgenic strains with UAS-ref(2)P were generated by P-element-mediated transformation. Transgenic flies were generated at BestGene Inc. (Chino Hills, CA, USA). At least eight independent transgenic lines were obtained and all were tested for protein expression. The pink1B9, park25, UAS-parkc2 and da-GAL4 flies were a gift from A. Withworth (MRC, Centre for Developmental and Biomedical Genetics, University of Sheffield). The atg1Δ3d line28 was kindly provided by Dr. T.P. Neufeld (University of Minnesota, Minneapolis, MN, USA). The ref(2)Pod2 and ref(2)Pod3 lines7 were described by Wyers et al.8 and were a kind gift from Dr. I. Nezis (Institute for Cancer Research, Oslo, Norway). Fly stocks and crosses were maintained on standard cornmeal agar media at 25 °C.
Molecular biology
Expression constructs encoding Drosophila Ref(2)P fused to an N-terminal HA tag sequence were generated by polymerase chain reaction (PCR) using appropriate adaptor primers, and the PCR fragments were cloned into the GAL4-responsive pUAST expression vector for the generation of transgenic fly strains.
Quantification of mitochondrial proteins
The concentration of Cyt c was determined using the Cytochrome c ELISA Kit (Invitrogen, Paisley, UK), and the activity of CSwas calculated using the Citrate Synthase Assay Kit (Sigma-Aldrich, Dorset, UK). Absorbances were measured on an Infinite 200 PRO multimode plate reader equipped with a Quad4 monochromator (TECAN, Mannedorf, Switzerland).
PCR amplification of mtDNA
Analysis of the mtDNA content was performed using quantitative real-time PCR on an Mx4000 (Stratagene, Santa Clara, CA, USA) real-time cycler using QuantiTect SYBR Green PCR system (QIAGEN, Manchester, UK). Specific primers for mtDNA (Forward primer: 5′-CAACCATTCATTCCAGCCTT-3′; Rev primer: 5′-GAAAATTTTAAATGGCCGCA-3′) were designed using the primer-BLAST tool (NCBI). The levels of mtDNA were normalised to the levels of nuclear DNA (nDNA) amplified using primers for dmActin (Dm_Act79B, QuantiTect Primer Assay, Cat. No. QT00967393). Quantification was performed using the comparative Ct method.29
To produce amplicons that cover the mtDNA genome 10 regions were amplified with the help of PCR, using the following primer sets: Set 1 (Forward primer: 5′-GGTTAGGAGCTTGAATAGG-3′, Reverse primer: 5′-CGCGATTATTGATTAAGTGGC-3′), Set 2 (Forward primer: 5′-GGAGGATTACCTCCATTTTTAGG-3′, Reverse primer: 5′-GTAATAGCTCCTGCTAGTACTGG-3′), Set 3 (Forward primer: 5′-TGGAGCTGGAACAGGATGAACTG-3′, Reverse primer: 5′-GTTGATCCAATAGTTGATAC-3′), Set 4 (Forward primer: 5′-TTGGCTGGAATACCTCGACG-3′, Reverse primer: 5′-CCATAAAATAAACCCGGTCG-3′), Set 5 (Forward primer: 5′-GAACAGTACCTGCTTTAGGAG-3′, Reverse primer: 5′-GCAGTTAATCGGACAGCTAATGT-3′), Set 6 (Forward primer: 5′-GCTCATTTAGTTCCTCAAGG-3′, Reverse primer: 5′-CATGCAGCTGCTTCAAAACC-3′), Set 7 (Forward primer: 5′-GCTCCATTTACTATTGCAGACTC-3′, Reverse primer: 5′-TACAAAATGAATTTGAAATG-3′), Set 8 (Forward primer: 5′-GAGCAACAGATGAATAAGC-3′, Reverse primer: 5′-GCTGGTTGGTCGTCTAATTC-3′), Set 9 (Forward primer: 5′-CCAGCTCCAATTAATATTTC-3′, Reverse primer: 5′-GCCGCAGTATTTTGACTGTG-3′), Set 10 (Forward primer: 5′-CGTCCAACCATTCATTCCAGC-3′, Reverse primer: 5′-CTAAATTGGTGCCAGCAGTCGCGG-3′). The PCR-generated amplicons partially overlap and cover 14 413 bp of the 19 501 bp Drosophila mitochondrial genome (73.9%). PCR amplification was performed using the New England Biolabs (Hitchin, UK) MasterMix Taq, and the length and quality of products verified using gel electrophoresis. PCR products were subsequently purified using the PCR purification kit (QIAGEN) and were pooled together.
Direct estimation of heteroplasmic mtDNA mutations using NGS
The estimation of DNA mutations was carried out using ion semiconductor sequencing on an Ion Torrent Personal Genome Machine (Ion PGM System).
mtDNA from total cellular DNA was enriched using a PCR-based approach (see above). Pooled PCR products were used for DNA library preparation using the Ion Xpress Plus Fragment Library Kit (Life Technologies Ltd, Paisley, UK). Adaptor-ligated products were then size-selected by gel purification and amplified to obtain genomic DNA libraries that were suitable for sequencing on the Ion PGM System. Sequencing reads with a mean read length of 90 bp were aligned to the reference Drosophila mitochondrial genome (release 5.49) using the Torrent Server Aligner plug-in (v3.6.56201, using the TMAP alignment algorithm). Mutations were detected using the Torrent Server variantCaller plug-in (v3.6.59049). Variants present in excess 1.6% per base were selected to account for PCR-associated errors.30
Immunofluorescence and confocal microscopy
To analyse the mitochondria and mtDNA nucleoids in the indirect flight muscles, adult fly thoraces were dissected, fixed in 4% paraformaldehyde and incubated with Hoechst 33342 or PicoGreen (Life Technologies Ltd), respectively. The indirect muscle fibres were then isolated and imaged on a Zeiss LSM510 confocal microscope (Zeiss, Cambridge, UK). For the analysis of dopaminergic neurons, fly brains were dissected from 20-day-old flies and stained for antityrosine hydroxylase (Immunostar Inc., Hudson, WI, USA). Brains were mounted on Vectashield (Vector Laboratories, Burlingame, CA, USA), imaged using confocal microscopy and TH-positive PPL1 cluster neurons were counted.
Statistical analysis
Data are presented as mean values, and error bars indicate ±S.D. or ±S.E.M. as noted. Inferential statistical analysis was performed using the Prism and StatMate software packages (www.graphpad.com). The significance level is indicated as *** for P<0.001, ** for P<0.01, * for P<0.05 and NS for P>0.05.
Locomotor analysis
Climbing assays were performed as previously described,31 using a countercurrent apparatus developed initially for phototaxis experiments. Twenty male flies (3––5 days old) were placed into the first chamber, taped to the bottom, then given 20 s to climb a distance of 10 cm. Flies that successfully climbed 10 cm or beyond in 20 s were then shifted to a new chamber, and both sets of flies were given another opportunity to climb the 10-cm distance. This procedure was repeated a total of five times. After five trials, the number of flies in each chamber was counted. At least 60 flies were used for each genotype tested.
Longevity assays
Groups of 10 newly eclosed males of each genotype were placed into separate vials with food and maintained at 25 °C. Flies were transferred to vials containing fresh food every 2–3 days, and the number of dead flies was recorded. The data are presented as Kaplan–Meier survival distributions, and the significance was determined by log-rank tests.
Electron microscopy
For TEM, adult fly thoraces or testes were fixed overnight in 0.1 M sodium cacodylate buffer (pH 7.4) containing 2% paraformaldehyde, 2.5% glutaraldehyde and 0.1% Tween-20. Samples were post-fixed for 1 h at room temperature in a solution containing 1% osmium tetroxide and 1% potassium ferrocyanide. After fixation, samples were stained en bloc with 5% aqueous uranyl acetate overnight at room temperature; the samples were then dehydrated via a series of ethanol washes and embedded in Taab epoxy resin (Taab Laboratories Equipment Ltd., Aldermaston, UK). Semithin sections were stained with toluidine blue and areas of the sections were selected for ultramicrotomy. Ultrathin sections were stained with lead citrate and recorded using a Megaview 3 digital camera and iTEM software (Olympus Soft Imaging Solutions GmbH, Münster, Germany) in a Jeol 100-CXII electron microscope (Jeol UK Ltd., Welwyn Garden City, UK).
For scanning electron microscopy (SEM), intact adult flies were initially processed as for TEM but, following dehydration, samples were dried with hexamethyldisilazane, mounted with silver DAG and sputter-coated with gold. Samples were recorded in the same microscope, using an ASID4DS scanning unit and IScan 2000 software (ISS Group, Manchester, UK). The orientation (anterior to the top), contrast and brightness of the images were adjusted using Adobe Photoshop CS3.
Preparation of Drosophila protein lysates and western blot analysis
Single adult flies (3 days old) were homogenised using a motorised pestle in lysis buffer (0.1% Triton-X100, 10 mM EDTA, 1 mM DTT, 100 mM KCl, 20 mM HEPES (pH 7.5), 5% Glycerol, 1 μg/ml chymostatin, 1 μg/ml leupeptin, 1 μg/ml pepstatin A and 1 mM PMSF). Lysates were subsequently cleared using centrifugation. Proteins were separated on 12% SDS-PAGE gels and transferred to Immobilon PVDF membranes (Millipore, Walford, UK). The membranes were blocked in 5% milk and then incubated with the indicated primary antibody (see Supplementary Table I) prior to incubation with the appropriate HRP-conjugated secondary antibody. Antibody complexes were visualised using enhanced chemiluminescence (ECL). Results are representative of at least three biological replicates per genotype.
Respirometry analysis
Mitochondrial respiration was assayed at 37 °C by high-resolution respirometry using an OROBOROS Oxygraph. The DatLab software package (OROBOROS, Innsbruck, Austria) was used for data acquisition (2-s time intervals) and analysis, including calculation of the time derivative of oxygen concentration, signal deconvolution dependent on the response time of the oxygen sensor and correction for instrumental background oxygen flux. Respiration was assayed by homogenising flies using a pestle in MiR05 respiration buffer (20 mM HEPES, 10 mM KH2PO4, 110 mM sucrose, 20 mM taurine, 60 mM K-lactobionate, 0.5 mM EGTA, 3 mM MgCl2, 1 g/l fatty-acid-free BSA). Complex I activity was assayed in MiR05 respiration buffer in the presence of 2 mM malate, 10 mM glutamate and 5–10 mM ADP. Complex II was assayed in respiration buffer supplemented with 1 mM rotenone, 10 mM succinate and 5–10 mM ADP. ADP was not added for measurement of respiration in uncoupled mitochondria obtained by one freeze (liquid nitrogen)–thaw cycle of fly tissue.
Abbreviations
- PD:
-
Parkinson’s disease
- ROS:
-
Reactive oxygen species
References
de Castro IP, Martins LM, Tufi R . Mitochondrial quality control and neurological disease: an emerging connection. Expert Rev Mol Med 2010; 12: e12.
Narendra D, Tanaka A, Suen DF, Youle RJ . Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 2008; 183: 795–803.
Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 2010; 12: 119–131.
Ziviani E, Tao RN, Whitworth AJ . Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci USA 2010; 107: 5018–5023.
Kirkin V, McEwan DG, Novak I, Dikic I . A role for ubiquitin in selective autophagy. Mol Cell 2009; 34: 259–269.
Pimenta de Castro I, Costa AC, Lam D, Tufi R, Fedele V, Moisoi N et al. Genetic analysis of mitochondrial protein misfolding in Drosophila melanogaster. Cell Death Differ 2012; 19: 1308–1316.
Nezis IP, Simonsen A, Sagona AP, Finley K, Gaumer S, Contamine D et al. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol 2008; 180: 1065–1071.
Wyers F, Petitjean AM, Dru P, Gay P, Contamine D . Localization of domains within the Drosophila Ref(2)P protein involved in the intracellular control of sigma rhabdovirus multiplication. J Virol 1995; 69: 4463–4470.
Magwere T, Goodall S, Skepper J, Mair W, Brand MD, Partridge L . The effect of dietary restriction on mitochondrial protein density and flight muscle mitochondrial morphology in Drosophila. J Gerontol A Biol Sci Med Sci 2006; 61: 36–47.
Legros F, Malka F, Frachon P, Lombes A, Rojo M . Organization and dynamics of human mitochondrial DNA. J Cell Sci 2004; 117: 2653–2662.
Berry C, La Vecchia C, Nicotera P . Paraquat and Parkinson's disease. Cell Death Differ 2010; 17: 1115–1125.
Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 2006; 441: 1162–1166.
Park J, Lee SB, Lee S, Kim Y, Song S, Kim S et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 2006; 441: 1157–1161.
Brand AH, Perrimon N . Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993; 118: 401–415.
Verstreken P, Ly CV, Venken KJ, Koh TW, Zhou Y, Bellen HJ . Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron 2005; 47: 365–378.
Tain LS, Mortiboys H, Tao RN, Ziviani E, Bandmann O, Whitworth AJ . Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat Neurosci 2009; 12: 1129–1135.
de Castro IP, Martins LM, Loh SH . Mitochondrial quality control and Parkinson's disease: a pathway unfolds. Mol Neurobiol 2011; 43: 80–86.
Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y . Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 2000; 150: 1507–1513.
Levine B, Klionsky DJ . Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 2004; 6: 463–477.
Matsuura A, Tsukada M, Wada Y, Ohsumi Y . Apg1p a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 1997; 192: 245–250.
Scott RC, Schuldiner O, Neufeld TP . Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell 2004; 7: 167–178.
Tatsuta T, Langer T . Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J 2008; 27: 306–314.
Reeve AK, Krishnan KJ, Turnbull D . Mitochondrial DNA mutations in disease, aging, and neurodegeneration. Ann N Y Acad Sci 2008; 1147: 21–29.
Kamel F . Epidemiology. Paths from pesticides to Parkinson's. Science 2013; 341: 722–723.
Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 2010; 8: e1000298.
Okatsu K, Saisho K, Shimanuki M, Nakada K, Shitara H, Sou YS et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells 2010; 15: 887–900.
Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007; 131: 1149–1163.
Scott RC, Schuldiner O, Neufeld TP . Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell 2004; 7: 167–178.
Schmittgen TD, Livak KJ . Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008; 3: 1101–1108.
He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE et al. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature 2010; 464: 610–614.
Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ . Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci USA 2003; 100: 4078–4083.
Acknowledgements
We would like to thank Alex Whitworth, Thomas Neufeld, Ioannis Nezis and the Bloomington Drosophila Stock Center for fly stocks; Judy McWilliam and Tim Smith for assistance with Electron Microscopy; Carolyn Jones for Next-Generation Sequencing assistance and Jalpa Parmar for fly food preparation. This work is funded by the Medical Research Council, UK. Inês Castro is recipient of a Ph.D. grant (SFRH/BD/41682/2007) from Fundação para a Ciência e a Tecnologia (FCT), Portugal.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Stephanou
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
de Castro, I., Costa, A., Celardo, I. et al. Drosophila ref(2)P is required for the parkin-mediated suppression of mitochondrial dysfunction in pink1 mutants. Cell Death Dis 4, e873 (2013). https://doi.org/10.1038/cddis.2013.394
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.394
Keywords
This article is cited by
-
Repeat length of C9orf72-associated glycine–alanine polypeptides affects their toxicity
Acta Neuropathologica Communications (2023)
-
Mouse models of SMA show divergent patterns of neuronal vulnerability and resilience
Skeletal Muscle (2022)
-
Pantothenate kinase 2 interacts with PINK1 to regulate mitochondrial quality control via acetyl-CoA metabolism
Nature Communications (2022)
-
Inhibition of Ref(2)P, the Drosophila homologue of the p62/SQSTM1 gene, increases lifespan and leads to a decline in motor function
BMC Research Notes (2021)
-
The autophagy receptor p62/SQST-1 promotes proteostasis and longevity in C. elegans by inducing autophagy
Nature Communications (2019)