Abstract
Oroxylin A is a major active component of the Chinese traditional medicinal plant Scutellaria baicalensis Georgi, which has been reported as a potential anticancer drug. We demonstrated that, Oroxylin A inhibited the glycolysis and the binding of hexokinase II (HK II) with mitochondria in human breast carcinoma cell lines, which was dependent on sirtuin-3 (SIRT3). The level of SIRT3 in mitochondria was increased by Oroxylin A. Then SIRT3 deacetylated cyclophilin D, diminished its peptidyl-prolyl cis-trans isomerase activity and induced its dissociation from the adenine nucleotide translocator. Finally, SIRT3-induced inactivation of cyclophilin D resulted in the detachment of mitochondrial HK II and the inhibition of glycolysis. These results have important implications for the metabolism reprogramming effect and the susceptibility to Oroxylin A-induced mitochondrial cytotoxicity through the regulation of SIRT3 in breast carcinoma.
Similar content being viewed by others
Main
Breast cancer comprises 22.9% of all cancers in women, which cause over 40 000 women (13.7% of cancer deaths in women) dying from the disease worldwide.1, 2 Traditionally, breast cancer is usually treated with surgery and then possibly with chemotherapy or radiation, or both. The prognosis of breast cancer is dependent on many prognostic factors, including tumor size, nodal status and histological grade.3 Compared with the well-defined behavior of breast cancer in a population, the molecular mechanism of breast cancer initiation and progression is still not studied deeply. Recently, it has been reported that NAD+-dependent protein deacetylation may have a role in breast cancer pathogenesis.4
Sirtuin (SIRT) proteins, class-III NAD+-dependent histone deacetylases (HDACs), are a druggable class of enzymes, which are involved with a variety of functions, including aging and regulation of metabolism,5, 6 and have beneficial effects on many human diseases.7 There are currently seven known SIRT family members, three of which (SIRT3, SIRT4, SIRT5) are localized to the mitochondria. SIRT3, which is the best studied of the mitochondrial SIRT proteins, has been shown to deacetylate and activate acetyl-CoA synthetase 2 (ACS2) and glutamate dehydrogenase (GDH), resulting in the increase of the tricarboxylic acid cycle (TAC) directly.8, 9 Moreover, SITR3 results in the considerable deacetylation of mitochondrial proteins, along with a decrease of glycolysis and a decrease in ATP levels under stress conditions.10
In the 1920s, Warburg11 shed the first light on the unique energy metabolism in cancer cell, who suggested a shift in energy production from mitochondrial oxidative phosphorylation (OXPHOS) to aerobic glycolysis. Aerobic glycolysis not only supports rapid growth, but also makes the cancer cell less dependent on oxygen availability and generates a suitable micro-environment. Inhibition of glycolysis has been a novel therapeutic focus in cancer therapies.12, 13 Regulation on the enzymes involved in glucose or lactate metabolism, such as hexokinase II (HK II), is an effective manner to inhibit glycolysis.14
The glycolysis of cancer cell metabolism is associated with the binding of HK II to the outer mitochondrial membrane protein voltage-dependent anion channel (VDAC).15 The increased binding of HK II induces mitochondria-mediated apoptosis and is associated with the increased resistance of cancer cells to chemotherapeutic drugs.16, 17 Importantly, cyclophilin D (also known as peptidyl-prolyl cis-trans isomerase D or PPID), which localizes to the mitochondrial matrix, is also necessary to promote the binding of HK II to VDAC.
Oroxylin A is a flavonoid isolated from Scutellaria root that exhibits multiple pharmacological activities, including anti-oxidative, anti-inflammatory, anti-viral and anti-tumor properties. Oroxylin A has been previously demonstrated to be a competitive candidate of novel anticancer drug in several types of cancers. Oroxylin A has multi-mechanism of anticancer, including apoptosis induction,18 metastasis inhibition,19 cell-cycle arrest induction,20 and so on. This study for the first time investigated the potential mechanism of Oroxylin A on glycolysis inhibition by modulating SIRT3.
Results
Oroxylin A inhibits glycolysis and stimulates the release of HK II from the mitochondria in breast carcinoma
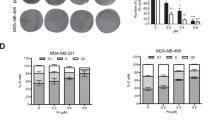
In the studies, clotrimazole (CTZ) was used as a positive control, which preferentially inhibited human breast cancer cells glycolysis and detached HK from mitochondria.21, 22 In MDA-MB-231 and MCF-7 cells, 100 μ M CTZ for 24 h, decreased the glucose uptake, lactate production, ATP generation and mitochondrial membrane potential (MMP), significantly (Figure 1). When treated with Oroxylin A, MDA-MB-231 and MCF-7 cells displayed significant decrease in glucose uptake and ATP generation in a concentration- and time-dependent manner (Figures 1a–d). As well the lactate generations in MDA-MB-231 and MCF-7 cells were decreased by 20% and 16.4%, respectively, upon 200 μ M Oroxylin A treatment for 48 h (Figures 1e and f) . Besides, the intracellular lactate content was decreased by two to threefold after Oroxylin A treatment in MDA-MB-231 and MCF-7 cells (Supplementary Figure 1A). Moreover, the expressions and the activities of some key glycolytic enzymes/factors, including glucose transporter-1 (Glut-1), 6-phosphofructo-2-kinase (PFK), lactate dehydrogenase (LDH) and pyruvate kinase (PK), were detected. As shown in Supplementary Figure 1B, 100 μ M CTZ decreased the expressions of PFK (PFKFB1/4 and PFKFB2 are the subtype of PFK) and PKM2, and 150 μ M Oroxylin A decreased the expressions of PKM2 and Glut-1. Both 100 μ M CTZ and 150 μ M Oroxylin A decreased the activity of HK, PFK, PK and LDH, significantly (Supplementary Figures 1D–G). These results suggested that the Oroxylin A inhibited the glycolysis in breast cancer cells. Moreover, the decrease of glycolysis by Oroxylin A for 48 h was accompanied by a decrease in MMP (Figure 1g). Oroxylin A inhibited the glycolysis, as well as the growth of MDA-MB-231 and MCF-7 cells (Figure 1h).
Oroxylin A inhibited the glycolysis in breast carcinoma. MDA-MB-231 and MCF-7 were treated with Oroxylin A upon different concentrations for 48 h, or upon 200 μ M Oroxylin A at various time, upon 100 μ M CTZ for 24 h. (a, b) Glucose uptake was measured using the Amplex Red assay. (c, d) Production of lactic acid was assayed by Lactic Acid production Detection kit. (e, f) Quantification of ATP generation was detected by the luminometer Orion II. The ratios of glucose uptake, produced lactic acid and ATP generation in treated groups compared with non-treated group are represented by histograms. (g) Cells were treated with Oroxylin A for 48 h. The loss of MMP was analyzed by JC-1 assay using flow cytometry. The percentages of the loss of MMP are represented by histograms. (h) Cells were treated with Oroxylin A for 48 h and cell viability was determined by MTT assay. Bars, S.D., *P<0.05 or **P<0.01 versus non-treated control of MDA-MB-231 cells, #P<0.05 or ##P<0.01 versus non-treated control of MCF-7 cells.
Many cancer cells display a great increasein binding of HK II to the mitochondria, which provides a metabolic and survival benefit.15, 23, 24 In previous studies, we have found that Oroxylin A could induce the dissociation of HK II from the mitochondria and inhibit glycolysis in A549 cells. Therefore, we wanted to determine whether the decrease of glycolysis by Oroxylin A had any relationship on the expression or localization of HK II in breast cancer cells. CTZ inhibited the detachment of HK from mitochondria (Figure 2). Oroxylin A caused a marked redistribution of HK II from the cytosol to the mitochondria both in MDA-MB-231 cells and MCF-7 cells. Importantly, besides the redistribution of HK II from the mitochondria to the cytosol, Oroxylin A also decreased the level of HK II expression (Figure 2a). Then we investigated the binding of HK II by immunoprecipitates. The binding capacity of HK II with VDAC diminished in a concentration-dependent manner, when cells were treated with Oroxylin A for 48 h (Figure 2b).
Oroxylin A suppressed HK II binding to VDAC in mitochondria. MDA-MB-231 and MCF-7 were treated with Oroxylin A (0, 100,150 and 200 μ M) for 48 h, or 100 μ M CTZ for 24 h. (a) Mitochondrial and cytosolic fractions were isolated after treatment and subjected to western blot analysis for HK II. (b) Mitochondria were isolated and HK II was immunoprecipitated using VDAC antibody. Western blot assays were performed for HK II and VDAC
The Oroxylin A-induced inhibition of glycolysis and HK II detachment from mitochondria is SIRT3 dependent
It has been reported that SIRT3 can regulate HK II binding with mitochondria.25 The loss-of-function in SIRT3 results in a damage-permissive and tumorigenic cellular environment. As shown in Figures 3a and b, compared with normal cell lines, SIRT3 was downregulated in many human cancer cell lines (including hepatoma, breast cancer, lung cancer and colon carcinoma) at the transcription and the post-transcription level.
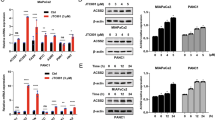
Oroxylin A stimulated SIRT3 expression and induced SIRT3-dependent release of HK II from the mitochondria. (a,b) Expression of SIRT3 proteins (a) and mRNA (b) in kinds of human cancer cell lines and human normal cell lines. (c) Cells were treated with Oroxylin A for 48 h. Mitochondrial fractions were isolated after treatment and subjected to western blot analysis for HK II. (d) MDA-MB-231 and MCF-7 cells were transfected with siRNA targeting SIRT3 or with a non-targeting control siRNA. Cells were then incubated with 200 μ M Oroxylin A for 48 h. After 48 h, the cells were harvested and mitochondria was isolated. Western blot assays were performed for HK II and SIRT3
Human SIRT3 is expressed as a full-length 44-kD protein that is targeted to the mitochondria by its N-terminal localization sequence.26 In the mitochondria, SIRT3 is cleaved via the mitochondrial matrix processing peptidase (MPP) to a short 28-kD protein, which is important for SIRT3 enzymatic activity.27 Dramatically, Oroxylin A upregulated the cleaved SIRT3 of the mitochondria in both breast cancer cells (Figure 3a). The total SIRT3 proteins in MCF-7 cells were also increased by Oroxylin A. To determine the roles SIRT3 played in the glycolysis inhibition and HK II detachment from mitochondria by Oroxylin A, siRNA was used to suppress the expression of SIRT3. As a result, suppression of SIRT3 expression largely prevented the Oroxylin A-induced decrease in glucose uptake, ATP production, lactate generation (Figure 4) and HK II detachment from mitochondria (Figure 3d). Moreover, transfection with siRNA targeting SIRT3 suppressed the decrease of HK II induced by Oroxylin A (Supplementary Figure 3B).
The inhibition of glycolysis by Oroxylin A was SIRT3 dependent. MDA-MB-231 and MCF-7 cells were transfected with siRNA targeting SIRT3 or with a non-targeting control siRNA. Cells were then incubated with 200 μ M Oroxylin A for 48 h. (a) Glucose uptake was measured using the Amplex Red assay. (b) Production of lactic acid was assayed by Lactic Acid production Detection kit. (c) Quantification of ATP generation was detected by the luminometer Orion II. The ratios of glucose uptake, produced lactic acid and ATP generation in treated groups compared with non-treated group are represented by histograms. Bars, S.D., *P<0.05 or **P<0.01 versus Oroxylin A-treated without siRNA group of MDA-MB-231 cells, #P<0.05 versus without siRNA group of MCF-7 cells
It is suggested that the Oroxylin A-induced inhibition of glycolysis and HK II detachment from mitochondria were SIRT3 dependent.
Oroxylin A stimulates full-length SIRT3 to translocate to the mitochondria from the nucleus upon cellular oxidative stress
In previous studies, we found that Oroxylin A increased SIRT3 in mitochondria. As shown in Figure 5a, the cleaved form of SIRT3 in mitochondria was increased. It has been reported that SIRT3 is transported from the nucleus to the mitochondria upon cellular stress.28 After the treatment of Oroxylin A for 48 h, the reactive oxygen level (ROS) was increased in a concentration- and time-dependent manner (Figure 5b). Then we investigated whether the increased ROS level by Oroxylin A was associated with the translocation of SIRT3. As shown in Figure 5c, SIRT3 was translocated from the nucleus to the mitochondria upon 200 μ M Oroxylin A or 1 mM H2O2. SIRT3 in mitochondria was also increased by Oroxylin A or H2O2. Moreover, NAC reversed the Oroxylin A-induced increase on SIRT3 in mitochondria (Figure 5d).
SIRT3 was expelled from the nucleus to the mitochondria by Oroxylin A upon cellular oxidative stress. (a) MDA-MB-231 cells were treated with Oroxylin A for 48 h. Mitochondrial and nuclear fractions were isolated after treatment and subjected to western blot analysis for full-length band and cleaved band of SIRT3. (b) MDA-MB-231 and MCF-7 cells were treated with Oroxylin A upon different concentrations or time. Quantification of ROS was detected using fluorescent dye DCFH-DA by FACSCalibur flow cytometry at Ex/Em of 488 nm/525 nm. Bars, S.D., *P<0.05 or **P<0.01 versus Oroxylin A-treated without siRNA group of MDA-MB-231 cells, #P<0.05 or ##P<0.01 versus without siRNA group of MCF-7 cells. (c) Cells were treated with 1 mM H2O2 for 36 h or 200 Oroxylin A for 48 h, respectively. Immunofluorescence experiment performed in MDA-MB-231 and MCF-7 cells upon oxidative stress or Oroxylin A treatment using antibodies specific to full-length of and cleaved SIRT3, DAPI and Mitotracker. (d) MDA-MB-231 cells were pretreated with10 mM NAC for 1 h, then treated with 1 mM H2O2 for 36 h or 200 μ M Oroxylin A for 48 h, respectively. Mitochondrial fractions were isolated after treatment and subjected to western blot analysis for HK II and SIRT3
These results suggested that the cellular oxidative stress caused by Oroxylin A could stimulate SIRT3 to translocate to the mitochondria.
Oroxylin A deacetylates and inactivates cyclophilin D by SIRT3
Several recent studies have reported that mitochondrial binding of HK II is affected by the activity of cyclophilin D.29, 30 Cyclophilin D is an immunophilin that exhibits peptidyl-prolyl cis-trans isomerase (PPIase) activity and is localized to the mitochondrial matrix. Therefore, cyclophilin D was immunoprecipitated and its acetylation status was determined with anti-acetylated-lysine antibodies. As shown in Figure 6a, cyclophilin D was deacetylated in Oroxylin A-treated MDA-MB-231 and MCF-7 cells. However, nicotinamide (NAM), which is a well-established potent inhibitor of the SIRT family of histone/protein deacetylases,31 blocked the deacetylation of cyclophilin D caused by Oroxylin A. Moreover, transfection with siRNA targeting SIRT3 suppressed the deacetylation of cyclophilin D induced by Oroxylin A as well (Figure 6d). It was suggested that Oroxylin A deacetylated cyclophilin D through SIRT3.
Oroxylin A-induced SIRT3-mediated deacetylation and inhibited the peptidyl-prolyl cis-trans isomerase activity of cyclophilin D, preventing the binding of cyclophilin D to ANT. (a–c) MDA-MB-231 and MCF-7 cells were treated with 200 μ M Oroxylin A and 10 Mm NAM, respectively, or both. (a) Mitochondria were isolated and acetylated cyclophilin D (acetylated Cyp-D) was immunoprecipitated using cyclophilin D antibody. Western blot assays were performed for acetylated-lysine and cyclophilin D(Cyp-D). (b) Mitochondria were isolated and ANT-1 was immunoprecipitated using cyclophilin D antibody. Western blot assays were performed for ANT-1 and cyclophilin D (Cyp-D). (c) Cyclophilin D was immunoprecipitated from mitochondrial lysates and its peptidyl-prolyl cis-trans isomerase activity determined. Bars, S.D.(±) (d, e) MDA-MB-231 and MCF-7 cells were transfected with siRNA targeting SIRT3 or with a non-targeting control siRNA. Cells were then incubated with 150 μ M Oroxylin A for 48 h. Mitochondria were isolated, then deacetylated cyclophilin D and ANT-1 was immunoprecipitated as above, respectively
Cyclophilin D possesses PPIase activity. Figure 6c shows that the activity of cyclophilin D was diminished in Oroxylin A-treated MDA-MB-231 and MCF-7 cells. While NAM reversed, the Oroxylin A decreased the activity of cyclophilin D. Thereby SIRT3-induced deacetylation of cyclophilin D could decrease the enzymatic activity of cyclophilin D.
Oroxylin A induces SIRT3-mediated dissociation of cyclophilin D from the adenine nucleotide translocator (ANT)
Cyclophilin D is thought to be localized exclusively to the mitochondrial matrix, modulating HK II binding to VDAC. A promising candidate for this intermediation role is the ANT, which is a component of the mitochondria permeability transition pore (PTP) and exert its pore-forming function by interacting with VDAC at mitochondrial contact sites.32
Cyclophilin D was coimmunoprecipitated with ANT-1 protein upon Oroxylin A for 48 h. As shown in Figure 6b, the binding of cyclophilin D to ANT-1 in Oroxylin A-treated MDA-MB-231 and MCF-7 cells were inhibited. However, when the cells were treated with Oroxylin A and NAM at the same time, there was a progressive decrease in the inhibition of cyclophilin D binding with ANT-1. The binding of ANT and cyclophilin D was also prevented by transfection with SIRT3 siRNA (Figure 6e). These results suggested that the Oroxylin A could inhibit the binding of ANT-1 with cyclophilin D in mitochondria, and SIRT3-mediated deacetylation of cyclophilin D was necessary for the interaction between cyclophilin D and ANT-1.
Deacetylation of cyclophilin D induced by Oroxylin A is necessary for dissociation of HK II from the mitochondria and inhibition of glycolysis
It has been reported that cyclophilin D stabilizes the interaction between ANT and VDAC, which is enhanced by HK II binding.33 To investigate the roles cyclophilin D played in the inhibition of glycolysis and the dissociation of HK II induced by Oroxylin A, wild-type cyclophilin D was overexpressed in MDA-MB-231 and MCF-7 cells.
As demonstrated in Figure 7e, overexpression of wild-type cyclophilin D did not reversed the inhibitory effects of Oroxylin A on the binding of HK II with VDAC. Moreover, the decrease of glucose uptake, ATP generation, lactate generation, as well as the loss of MMP induced by Oroxylin A, were not reversed by overexpression of cyclophilin D (Figures 7a–d). By contrast, NAM suppressed the decrease in glucose uptake, ATP generation, lactate generation and MMP caused by Oroxylin A. The inhibitory effects of Oroxylin A on the binding of HK II to the mitochondria was also reversed by NAM. It was suggested that Oroxylin A preferred to decreasing the activity of cyclophilin D via deacetylation, rather than changing the cyclophilin D expression. Deacetylation of cyclophilin D induced by Oroxylin A was necessary for dissociation of HK II from the mitochondria and inhibition of glycolysis.
The regulation on peptidyl-prolyl cis-trans isomerase activity of cyclophilin D was necessary for inhibition of glycolysis by Oroxylin A, instead of the expression of wide cyclophilin D. MDA-MB-231 and MCF-7 cells overexpressing wide cyclophilin D or treated with 10 mM NAM were incubated in the absence or presence of 150 μ M Oroxylin A for 48 h. (a–d) Glucose uptake, produced lactic acid, ATP generation and the loss of MMP were measured as previous. The ratios of glucose uptake (a), produced lactic acid (b) ATP generation (c) in treated groups compared with non-treated group, and the percentages of the loss of MMP (d) are represented by histograms. Bars, S.D., *P and #P<0.05 or **P and ##P<0.01. (e) Mitochondria were isolated and HK II was immunoprecipitated using VDAC antibody. Western blot assays were performed for HK II and VDAC
Discussion
Cancer cells undergo significant metabolic adaptation, which are much different from that in normal cells. Studies of the metabolic changes provide rationale and insight for anticancer therapy. The present study, for the first time, demonstrates that the natural flavanoid Oroxylin A influences the glycolysis to inhibit the growth of breast cancer, which is involved with the NDA+-dependent deacetylase SIRT3. As shown in Figure 8, Oroxylin A increased the expression, as well as the mitochondrial translocation of SIRT3, which resulted in the deacetylation of cyclophilin D and then provoked a dissociation of HK II from the mitochondria.
Oroxylin A-induced SIRT3-mediated cyclophilin D deacetylation, triggered HK II detachment, and inhibited glycolysis. Oroxylin A caused an increase in the expression and activity of SIRT3. In turn, SIRT3 promoted deacetylation of cyclophilin D (Cyp-D), resulting the dissociation of Cyp-D from ANT-1, and promoting the detachment of HK II from the mitochondria VDAC. As a result, the glycolysis was inhibited by Oroxylin A
According to Warburg, the bioenergetic switch from a higher energy yielding process (TCA cycle) to a lower energy yielding process (glycolysis) is a hallmark of most cancer cells.34 Although the aerobic glycolysis have significant roles in cancer metabolism, the function of OXPHOS is at the same position, or more important most of the time. It was reported that the predominant energy metabolism in breast cancer cells were glycolysis and OXPHOS.35 Therefore, we first investigate whether Oroxylin A influenced OXPHOS by measuring the oxygen consumption. As shown in Supplementary Figure 1C, Oroxylin A had a negligible influence on oxygen consumption compared with the control group. Thus, we focused on glycolysis. In addition to the enzymatic roles, some glycolytic enzymes, such as HK, have functions in transcriptional regulation, as well as apoptosis induction.36, 37 Therefore the use of glycolytic inhibitors, especially those targeting HK II, is a feasible approach for targeting cancer’s metabolic shortcomings.38 Several glycolytic inhibitors are being designed to target the glycolytic enzymes and synergy with other anticancer drugs. Methyl jasmonate is such a newly discovered ‘star’, which provided a new gateway in the cancer metabolic targeting. It has been reported that methyl jasmonate interfered with the interactions between HK and VDAC leading to the disruption of mitochondria.39 Previously, we have found that Oroxylin A could inhibit glycolysis by detaching HK II from VDAC in A549 cells. In present studies, Oroxylin A caused the dissociation of HK II from the mitochondria as well (Figure 2), and in turn inhibited the glycolysis (Figure 1 and Supplementary Figure 1). The activity of HK II was decreased by Oroxylin A as well Supplementary Figure 3A). Among the four rate-limiting enzymes, only the expression and activity both were decreased by Oroxylin A. Although Oroxylin A decreased the expression of Glut-1, the inhibitory degree on Glut-1 was weaker than on HK II. It was suggested that HK II, which was related with the regulation on SIRT3 (Figure 3c), played an important role in Oroxylin A-regulated glycolysis. Moreover, Oroxylin A treatment increased the apoptotic sensitivity of MCF-7/ADR cells to adriamycin (Supplementary Figure 3C).
Histone deacetylase Silent Information Regulator 2 (Sir2) homologs are a serious of NAD+-dependent protein deacetylases (Class 3 HDACs).40, 41 The biochemical activity of Sir2 orthologs is unique. NAD and NADH are involved in hundreds of metabolic reactions in cells. Thus, the biochemical activity of Sir2 led to the idea that SIRT is crucial for lifespan extension in response to metabolic and other environmental stressors.42, 43 The character that SIRT3 is the only member of the seven SIRTs linked genetically to lifespan in humans,44 together with the mitochondrial localization of SIRT3, made SIRT3 an especially interesting target for study. There is emerging evidence of a role for several SIRTs in carcinogenesis. Although SIRT3’s function varies in different normal and tumor tissues and may be cell- and tumor-type specific, our studies suggested SIRT3 played as a tumor suppressor in human breast cancer. In agreement with previous reports by Kim et al.,3, 10, 45, 46 we found SIRT3 was low-expressed in many human cancer cell lines than in normal cells (Figures 3a and b). Because of its localization to mitochondria, SIRT3 is required for the modulation of energy metabolism, apoptosis and ageing. We found the inhibitory effects of Oroxylin A on the glycolysis and HK II mitochondrial binding were dependent on SIRT3 (Figure 3d and Figure 4). In addition to SIRT3, the other two factors, AKT and GSK3β, were reported to regulate the binding of HK II to VDAC by phosphorylation.47, 48 As shown in Supplementary Figure 2A, Oroxylin A had little influence on the activation of AKT and GSK3β. After AKT was knockout, little changes happened to the HK II detachment from the mitochondria upon Oroxylin A treatment (Supplementary Figure 2B). Therefore, SIRT3 played an key role in the Oroxylin A-induced HK II detachment from VDAC.
Oroxylin A could increase the translocation of SIRT3 to mitochondria. SIRT3 is a nuclear NAD+-dependent HDAC that translocates to the mitochondria in response to various stressors, such as ultraviolet (UV) irradiation and the chemotherapeutic drug.28 We found that the increased translocation of SIRT3 by Oroxylin A was associated with the increased oxidative stress (Figures 5c and d). The reasons why Oroxylin A increased intracellular ROS level and oxidative stress, were attributed to the hydrogen-donating capacity of flavonoids, the influence of the mitochondrial function, as well as the regulation of the redox enzymes.18, 49, 50 Upon the cellular oxidative stress, which may be caused by overloading the cell with high expression of SIRT3, Oroxylin A would result in the expulsion of full-length SIRT3 from the nucleus. After reaching mitochondria, premature SIRT3 is cleaved at the N′-terminus targeting peptide to achieve its maximal deacetylase activity.51 It was reported that the mitochondrial import of SIRT3 is dependent on an NH 2-terminal amphipathic α-helix rich in basic residues.26
The mitochondrial SIRT3 regulates the activity of metabolic enzymes via protein deacetylation and/or mono-ADP-ribosylation. Unlike the numerous substrates of SIRT1,52 SIRT3 just has few specific substrates. This character provides a superiority to develop SIRT3 inhibitor with higher selectivity and lower side-effects. SIRT3 deacetylates cyclophilin D,25 diminishing its activity and inducing its dissociation from ANT. And cyclophilin D can stabilize the HK II mitochondrial binding.30 These findings suggest that SIRT3 have important roles in the mitochondrial functions and metabolism of cancer cells. Recent studies reported that SIRT3 can regulate mPTP through the deacetylation of cyclophilin D at lysine166, suppressing age-related cardiac hypertrophy.53 In our studies, Oroxylin A caused SIRT3-mediated deactylation of cyclophilin D (Figure 6d), decreased PPIase activity of cyclophilin D (Figure 6c), and resulted in the separation of cyclophilin D and ANT (Figure 6b). Additionally, the SIRT3-mediated deacetylation of cyclophilin D by Oroxylin A provoked a dissociation of HK II from the mitochondria (Figure 7e). When the SIRT3-mediated deacetylation of cyclophilin D was prevented, the decrease in glycolysis and mitochondrial dyfusion induced by Oroxylin A were reversed (Figures 7a–d). Interestingly, we found Oroxylin A affected the activity of cyclophilin D, rather than its expression. Because the overexpression of mutate cyclophilin D had little reversed effect on the Oroxylin A-induced inhibition of glycolysis. Instead, NAM could reverse the inhibitory effects of Oroxylin A by inhibiting SIRT3 activity.
Although several studies have implicated that SIRTs are involved in tumorigenesis, how SIRTs are involved in cancer is still not clear and is controversial. SIRT inhibitors/modifiers might have therapeutic benefit. Several inhibitors and activators of SIRTs have been tested in different cancer cell lines, but few have been tested in vivo.54 The SIRT inhibitors, NAM, induced apoptosis in lung cancer55 and oral cancer cells.46 In our studies, SIRT3 played as a tumor suppressor in human breast cancer. Although SIRT3 have important roles in tumor progression and life span, little specific modifiers for SIRT3 were explored and the majority of studies focused on human SIRT1 and/or SIRT2. Those drugs that are effective for several SIRT family members, may result some redundancy or even opposing actions unexpected. We have found that Oroxylin A increased the SIRT3 level in mitochondria and the effect of Oroxylin A on SIRT1 and SIRT2 is being investigated.
SIRT3 belong to the class-III HDAC. So far, class-I and II HDAC inhibitors have been tested in phase-I and II clinical trials. Some of the agents, such as trichostatin, were used to treat patients with solid tumors, hematologic malignancies and advanced leukemias.56 To our knowledge, there are no published reports on clinical trials using class-III HDAC inhibitors of SIRTs to treat cancer. Our studies suggested that Oroxylin A as class-III HDAC activators of SIRTs are especially potential. By upregulating SIRT3 level in mitochondria, Oroxylin A caused detachment of HK II from the mitochondria and inhibited the glycolysis in breast cancer. The reversal of metabolic reprogramming is an attractive strategy to increase susceptibility to therapy.
Materials and Methods
Reagents
Oroxylin A (C16H12O5) was isolated from the root of Scutellaria baicalensis according to previously reported protocols,57 dissolved in DMSO as a stock solution at −20 °C, and diluted with medium before each experiment. The final DMSO concentration did not exceed 0.1% throughout the study. Hydrogen peroxide (H2O2) solution (30%) was purchased from Sigma (Sigma-Aldrich, St. Louis, MO, USA) and diluted to 10−1M with distilled water. Nicotinamide (NAM) was purchased from Beyotime (Beyotime Institute of BioTechnology, Haimen, China) and diluted to 10−1M with distilled water. CTZ was purchased from Santa Cruz (Santa Cruz, CA, USA) and dissolved in DMSO.
Cell culture
The human breast cancer cell lines MDA-MB-231 cells and MCF-7 cells were purchased from Cell Bank of Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). MDA-MB-231 cells and MCF-7 cells, were respectively cultured in PRMI-1640 and Dulbecco’s Modified Eagle Medium (GIBCO, Invitrogen Corporation, Carlsbad, CA, USA) supplemented with 10% heat-inactivated fetal bovine serum (Sijiqing, Hangzhou, China), 100 U/ml penicillin G, and 100 μg/ml streptomycin at 37 °C with 5% CO2.
Cell viability inhibition assays
Cell viability was measured using the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) assay. Cells (1 × 104) were treated with Orxoylin A for 48 h at various concentrations (0–250 μ M). Absorbance of the resulting formazan was measured spectrophotometrically at 570 nm by the Universal Microplate Reader EL800 (BIO-TEK instruments, Inc., Vermont, MA, USA).
Lactic acid production
After treatment, to determine the generation of lactic acid, media was collected and assayed following the manufacturer’s instructions of Lactic Acid production Detection kit (KeyGen, Nanjing, China). The assay was detected by spectrophotometer (Thermo, Waltham, MA, USA) at 570 nm.
ATP assessment
Cellular ATP concentrations were measured with ATP Bioluminescent Somatic Cell Assay Kit obtained from Sigma (Sigma, St. Louis, MO, USA). After cells were incubated with Oroxylin A 48 h, cultured cells were lysed in an ice-cold ATP releasing buffer. Using an ATP standard provided by Sigma, ATP concentrations were then determined with the kit, and normalized to protein concentrations. Aliquots of 100 μl were transferred to white 96-well assay plates. Following addition of 100 μl luciferin and luciferase, luminescence was monitored on a luminometer Orion II (Berthold DS, Bleichstr, Pforzheim, Germany).
Glucose uptake assay
For the analysis of glucose uptake, the Amplex Red Glucose Assay Kit (Invitrogen, Eugene, OR, USA) was used. After treatment, media was collected and diluted 1 : 4,000 in water. The amount of glucose in the media was then detected using the Amplex Red Assay according to the manufacturer’s instructions. Glucose uptake was determined by subtracting the amount of glucose in each sample from the total amount of glucose in the media (without cells). The detection was performed by spectrophotometer (Thermo) at Ex/Em=530 nm/590 nm.
MMP determination
Following treatment, cells were harvested and resuspended with ice-cold PBS (2000 r.p.m. × 5 min). Quantitative changes of MMP at the early stage of cell apoptosis were measured by the Mitochondrial membrane potential Detection kit (KeyGen, Nanjing, China) with the lipophilic cation 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanine iodide (JC-1) and detected by FACSCalibur flow cytometry (Becton Dickinson, San Jose, CA, USA).
ROS measurement
ROS level was assessed using the fluorescent dye 2′7′-dichlorfluorescein-diacetate (DCFH-DA, Beyotime Institute of BioTechnology, Haimen, China) following the methods reported by Wei et al.49
Preparation of nuclear-, mitochondrial- and cytosol-enriched extracts
After cells were incubated with Oroxylin A for 48 h, Mitochondria/Cytosol Fractionation was collected following the instructions included with Mitochondria/Cytosol Fractionation Kit (BioVision, VA). Cell nuclear and cytoplasmic fractions were prepared using a nuclear/cytosol fractionation kit of Biovision Inc. (Mountain View, CA, USA) according to the manufacture’s direction.
Western blot
Samples were isolation with lysis buffer and eluted with SDS buffer, separated on SDS-polyacrylamide gels, and electroblotted onto PVDF membranes.58 Immunoreactive protein bands were detected using an Odyssey Scanning System (LI-COR Inc., Superior St., Lincoln, NE, USA). The following antibodies were used for western blot: VDAC, cyclophilin D, ANT (Santa Cruz) at 1 : 400 dilution; Sirt3, HK II, COX IV (Cell signaling Technology, Inc., Danvers, MA, USA) at 1 : 800 dilution; SIRT3 of nuclear protein was purchased from Abcam (Abcam Ltd, HK, China) at 1 : 1000 dilution.
Reverse transcription-PCR assay
Total cellular RNA was extracted from cells using TriPure solution (Roche Carolina, Florence, SC, USA) following the manufacturer’s instructions. The purity of the extracted RNA was determined by the ratio of A260/A280 using a BioPhotometer (Eppendorf, Hamburg, Germany). Reverse transcription-PCR was performed by PrimeScript reverse transcriptase (Takara Bio Inc., Otsu, Shiga, Japan) following the manufacturer’s instructions. The amplified PCR products were separated by electrophoresis on a 2% agarose gel containing ethidium bromide and quantitated by relative intensities of the bands as compared with those of β-actin using a GeneGenius BioImaging System with GeneSnap software (SYNGENE, Cambridge, UK). A value of 100% was given to the intensity of bands of untreated cells (control). The sequences of the PCR primers and the expected sizes of amplicons were as follows β-actin, 5′-CTGTCCCTGTATGCCTCTG-3′(sense) and 5′-ATGTCACGCACGAT-TTCC-3′ (antisense); SIRT3, 5′-CATTAAATGTGGTGGAACAAGAGGCCTG-3′ (sense) and 5′-AGTTCCTCTCCTTTGTAATCCCTCCGAC-3′ (antisense).
Immunoprecipitation
VDAC and ANT was immunocaptured from mitochondrial extracts using polyclonal antibodies to VDAC crosslinked to protein G-agarose beads (Santa Cruz). The immunocomplexes were analyzed by western blotting and probed with antibody against HK II.
ANT was immunocaptured from mitochondrial extracts using polyclonal antibodies to ANT crosslinked to protein G-agarose beads. The immunocomplexes were analyzed by western blotting and probed with antibody against cyclophilin D.
Cyclophilin D was immunocaptured from mitochondrial extracts using polyclonal antibodies to cyclophilin D crosslinked to protein G-agarose beads. The acetylated cyclophilin D were analyzed by western blotting and probed with acetylated-lysine antibody (Cell signaling Technology).
Immunofluorescence and confocal fluorescence microscopy
Cells were fixed with 4% paraformaldehyde in PBS at 1-h intervals, permeabilized with 0.5% Triton X-100, and blocked with 3% BSA for 30 min. Incubation with primary antibodies (diluted 1 : 50) (Abcam) against SIRT3 was done overnight at 4 °C. Mitochondrial were visualized with Mitotracker Red (Molecular Probes, Inc., Eugene, OR, USA), incubated at a final concentration of 150 nM for 45 h at 37 °C. Then the nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich) 20 min before imaging. A laser scanning confocal microscope FV10-ASW (Ver 2.1) (Olympus Corp, MPE FV1000, Tokyo, Japan) was used for colocalization analysis.
Measurement of cyclophilin D activity
Cyclophilin D protein was obtained from mitochondrial extracts. Cyclophilin D PPIase activity was determined colorimetrically using a peptide, in which the rate of conversion of cis to trans of a proline residue in the substrate peptide cis-Nsuccinyl-Ala-Ala-Pro-Phe-pnitroanilide (Sigma-Aldrich) makes it susceptible to cleavage by chymotrypsin, resulting in the release of the chromogenic dye, pnitroanilide. The absorbance change at 380 nm was monitored over a 5 min period with data collected every 20 s.
siRNA-mediated knockdown of SIRT3
The siRNAs targeting SIRT3 (Santa Cruz) were delivered by a lipid based method using Lipofectamine 2000 (Invitrogen Life Technologies, Grand Island, NY, USA) at a final siRNA concentration of 30 μ M. After formation of the siRNA–liposome complexes, the mixture was added to the breast cancer cells for 4 h. The medium was then aspirated, and complete medium containing 200 μ M Oroxylin A was added.
Transfection of wide cyclophilin D
Homo sapiens PPID human cDNA clone was obtained from OriGene (OriGene Technologies, Inc., Rockville, MD, USA). For transfection, cells were seeded in six-well plates at 65% confluency at first. Then the plasmid DNA (1 μg) was introduced into the cells using Lipofectamine 2000 (Invitrogen Life Technologies) according to the manufacturer’s recommendations. After that, cells were exposed to Oroxylin A or the vehicle and harvested for further experiments.
Statistical evaluation
Data are presented as mean ± S.D. from triplicate parallel experiments unless otherwise indicated. Statistical analyses were performed using one-way ANOVA.
Abbreviations
- SIRT3:
-
Sirtuin-3
- ACS2:
-
acetyl-CoA synthetase 2
- GDH:
-
glutamate dehydrogenase
- TAC:
-
tricarboxylic acid cycle
- OXPHOS:
-
oxidative phosphorylation
- HK II:
-
hexokinase II
- VDAC:
-
voltage-dependent anion channel
- NAM:
-
Nicotinamide
- MTT:
-
3-[4,5-dimethylthiazol-2-yl] -2,5-diphenyltetrazolium bromide
- MMP:
-
mitochondrial membrane potential
- DCFH-DA:
-
2′7′-dichlorfluorescein-diacetate
- DAPI:
-
4′,6-diamidino-2-phenylindole
- MPP:
-
matrix processing peptidase
- ROS:
-
reactive oxygen species
- PPIase:
-
peptidyl-prolyl cis-trans isomerase
- ANT:
-
adenine nucleotide translocator
- mPTP:
-
mitochondrial permeability transition pore
- Sir2:
-
Histone deacetylase Silent Information Regulator 2
- HDACs:
-
histone deacetylases
- UV:
-
ultraviolet
- CTZ:
-
Clotrimazole
- Glut-1:
-
glucose transporter-1
- PFK:
-
6-phosphofructo-2-kinase
- LDH:
-
lactate dehydrogenase
- PK:
-
pyruvate kinase
References
Gompel A, Santen RJ . Hormone therapy and breast cancer risk 10 years after the WHI. Climacteric 2012; 15: 241–249.
Smith RA, Duffy SW, Tabar L . Breast cancer screening: the evolving evidence. Oncology (Williston Park) 2012; 26: 471–475, 479–481, 485–486.
Ashraf N, Zino S, Macintyre A, Kingsmore D, Payne AP, George WD et al. Altered sirtuin expression is associated with node-positive breast cancer. Br J Cancer 2006; 95: 1056–1061.
Venugopal B, Evans TR . Developing histone deacetylase inhibitors as anti-cancer therapeutics. Curr Med Chem 2010; 18: 1658–1671.
Saunders LR, Verdin E . Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene 2007; 26: 5489–5504.
Schwer B, Verdin E . Conserved metabolic regulatory functions of sirtuins. Cell Metab 2008; 7: 104–112.
Westphal CH, Dipp MA, Guarente L . A therapeutic role for sirtuins in diseases of aging? Trends Biochem Sci 2007; 32: 555–560.
Hallows WC, Lee S, Denu JM . Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci USA 2006; 103: 10230–10235.
Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C . Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol 2008; 382: 790–801.
Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 2010; 17: 41–52.
Warburg O . On the origin of cancer cells. Science 1956; 123: 309–314.
Hamanaka RB, Chandel NS . Targeting glucose metabolism for cancer therapy. J Exp Med 2012; 209: 211–215.
Dang CV . Links between metabolism and cancer. Genes Dev 2012; 26: 877–890.
Wallace DC . Mitochondria and cancer: Warburg addressed. Cold Spring Harb Symp Quant Biol 2005; 70: 363–374.
Pedersen PL . Warburg, me and Hexokinase 2: multiple discoveries of key molecular events underlying one of cancers' most common phenotypes, the ‘Warburg Effect’, i.e., elevated glycolysis in the presence of oxygen. J Bioenerg Biomembr 2007; 39: 211–222.
Abu-Hamad S, Zaid H, Israelson A, Nahon E, Shoshan-Barmatz V . Hexokinase-I protection against apoptotic cell death is mediated via interaction with the voltage-dependent anion channel-1: mapping the site of binding. J Biol Chem 2008; 283: 13482–13490.
Shoshan-Barmatz V, Keinan N, Zaid H . Uncovering the role of VDAC in the regulation of cell life and death. J Bioenerg Biomembr 2008; 40: 183–191.
Liu W, Mu R, Nie FF, Yang Y, Wang J, Dai QS et al. MAC-related mitochondrial pathway in oroxylin-A-induced apoptosis in human hepatocellular carcinoma HepG2 cells. Cancer Lett 2009; 284: 198–207.
Lu Z, Lu N, Li C, Li F, Zhao K, Lin B et al. Oroxylin A inhibits matrix metalloproteinase-2/9 expression and activation by up-regulating tissue inhibitor of metalloproteinase-2 and suppressing the ERK1/2 signaling pathway. Toxicol Lett 2012; 209: 211–220.
Yang Y, Hu Y, Gu HY, Lu N, Liu W, Qi Q et al. Oroxylin A induces G2/M phase cell-cycle arrest via inhibiting Cdk7-mediated expression of Cdc2/p34 in human gastric carcinoma BGC-823 cells. J Pharm Pharmacol 2008; 60: 1459–1463.
Furtado CM, Marcondes MC, Sola-Penna M, de Souza ML, Zancan P . Clotrimazole preferentially inhibits human breast cancer cell proliferation, viability and glycolysis. PLOS One 2012; 7: e30462.
Penso J, Beitner R . Clotrimazole and bifonazole detach hexokinase from mitochondria of melanoma cells. Eur J Pharmacol 1998; 342: 113–117.
Pedersen PL, Mathupala S, Rempel A, Geschwind JF, Ko YH . Mitochondrial bound type II hexokinase: a key player in the growth and survival of many cancers and an ideal prospect for therapeutic intervention. Biochim Biophys Acta 2002; 1555: 14–20.
Pastorino JG, Hoek JB . Regulation of hexokinase binding to VDAC. J Bioenerg Biomembr 2008; 40: 171–182.
Shulga N, Wilson-Smith R, Pastorino JG . Sirtuin-3 deacetylation of cyclophilin D induces dissociation of hexokinase II from the mitochondria. J Cell Sci 2010; 123: 894–902.
Schwer B, North BJ, Frye RA, Ott M, Verdin E . The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol 2002; 158: 647–657.
Cooper HM, Huang JY, Verdin E, Spelbrink JN . A new splice variant of the mouse SIRT3 gene encodes the mitochondrial precursor protein. PLOS One 2009; 4: e4986.
Scher MB, Vaquero A, Reinberg D . SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev 2007; 21: 920–928.
Chiara F, Castellaro D, Marin O, Petronilli V, Brusilow WS, Juhaszova M et al. Hexokinase II detachment from mitochondria triggers apoptosis through the permeability transition pore independent of voltage-dependent anion channels. PLOS One 2008; 3: e1852.
Machida K, Ohta Y, Osada H . Suppression of apoptosis by cyclophilin D via stabilization of hexokinase II mitochondrial binding in cancer cells. J Biol Chem 2006; 281: 14314–14320.
Denu JM . Vitamin B3 and sirtuin function. Trends Biochem Sci 2005; 30: 479–483.
Vieira HL, Haouzi D, El Hamel C, Jacotot E, Belzacq AS, Brenner C et al. Permeabilization of the mitochondrial inner membrane during apoptosis: impact of the adenine nucleotide translocator. Cell Death Differ 2000; 7: 1146–1154.
Aniya Y, Imaizumi N . Mitochondrial glutathione transferases involving a new function for membrane permeability transition pore regulation. Drug Metab Rev 2011; 43: 292–299.
Pathania D, Millard M, Neamati N . Opportunities in discovery and delivery of anticancer drugs targeting mitochondria and cancer cell metabolism. Adv Drug Deliv Rev 2009; 61: 1250–1275.
Moreno-Sanchez R, Rodriguez-Enriquez S, Marin-Hernandez A, Saavedra E . Energy metabolism in tumor cells. FEBS J 2007; 274: 1393–1418.
Mathupala SP, Ko YH, Pedersen PL . Hexokinase II cancer’s double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene 2006; 25: 4777–4786.
Robey RB, Hay N . Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene 2006; 25: 4683–4696.
Rosano C . Molecular model of hexokinase binding to the outer mitochondrial membrane porin (VDAC1): implication for the design of new cancer therapies. Mitochondrion 2011; 11: 513–519.
Goldin N, Arzoine L, Heyfets A, Israelson A, Zaslavsky Z, Bravman T et al. Methyl jasmonate binds to and detaches mitochondria-bound hexokinase. Oncogene 2008; 27: 4636–4643.
Imai S, Armstrong CM, Kaeberlein M, Guarente L . Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 2000; 403: 795–800.
Blander G, Guarente L . The Sir2 family of protein deacetylases. Annu Rev Biochem 2004; 73: 417–435.
Gorospe M, de Cabo R . AsSIRTing the DNA damage response. Trends Cell Biol 2008; 18: 77–83.
Guarente L, Picard F . Calorie restriction—the SIR2 connection. Cell 2005; 120: 473–482.
Rose G, Dato S, Altomare K, Bellizzi D, Garasto S, Greco V et al. Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp Gerontol 2003; 38: 1065–1070.
Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell 2011; 19: 416–428.
Alhazzazi TY, Kamarajan P, Joo N, Huang JY, Verdin E, D'Silva NJ et al. Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer 2010; 117: 1670–1678.
Gottlob K, Majewski N, Kennedy S, Kandel E, Robey RB, Hay N . Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes Dev 2001; 15: 1406–1418.
Korzick DH, Kostyak JC, Hunter JC, Saupe KW . Local delivery of PKCepsilon-activating peptide mimics ischemic preconditioning in aged hearts through GSK-3beta but not F1-ATPase inactivation. Am J Physiol Heart Circ Physiol 2007; 293: H2056–H2063.
Wei L, Lu N, Dai Q, Rong J, Chen Y, Li Z et al. Different apoptotic effects of wogonin via induction of H(2)O(2) generation and Ca(2+) overload in malignant hepatoma and normal hepatic cells. J Cell Biochem 2010; 111: 1629–1641.
Kachadourian R, Day BJ . Flavonoid-induced glutathione depletion: potential implications for cancer treatment. Free Radic Biol Med 2006; 41: 65–76.
Pereira CV, Lebiedzinska M, Wieckowski MR, Oliveira PJ . Regulation and protection of mitochondrial physiology by sirtuins. Mitochondrion 2011; 12: 66–76.
Smith BC, Hallows WC, Denu JM . Mechanisms and molecular probes of sirtuins. Chem Biol 2008; 15: 1002–1013.
Hafner AV, Dai J, Gomes AP, Xiao CY, Palmeira CM, Rosenzweig A et al. Regulation of the mPTP by SIRT3-mediated deacetylation of CypD at lysine 166 suppresses age-related cardiac hypertrophy. Aging 2010; 2: 914–923.
Porcu M, Chiarugi A . The emerging therapeutic potential of sirtuin-interacting drugs: from cell death to lifespan extension. Trends Pharmacol Sci 2005; 26: 94–103.
Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 2001; 107: 137–148.
Acharya MR, Sparreboom A, Venitz J, Figg WD . Rational development of histone deacetylase inhibitors as anticancer agents: a review. Mol Pharmacol 2005; 68: 917–932.
Li HB, Chen F . Isolation and purification of baicalein, wogonin and oroxylin A from the medicinal plant Scutellaria baicalensis by high-speed counter-current chromatography. J Chromatogr A 2005; 1074: 107–110.
Zhao L, Guo QL, You QD, Wu ZQ, Gu HY . Gambogic acid induces apoptosis and regulates expressions of Bax and Bcl-2 protein in human gastric carcinoma MGC-803 cells. Biol Pharm Bull 2004; 27: 998–1003.
Acknowledgements
This work was supported by the National Science and Technology Major Project (No. 2012ZX09103101-050 and No.2012ZX09304-001), the Project Program of State Key Laboratory of Natural Medicines, China Pharmaceutical University (No. JKGZ201101 and SKLNMZZ201210), the National Natural Science Foundation of China (No. 30973556, No.91029744 and No. 81173086), Natural Science Foundation of Jiangsu Province, China (No. BK2010432), Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT-IRT1193), Graduate Innovation Fund of SIMCERE PHARMACEUTICAL GROUP, China (No. CX11B-006XS), Doctoral Talents Cultivation Plan of China Pharmaceutical University, China (No. 2011 II BPY07). Program for New Century Excellent Talents in University (NCET-12-0973).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Stephanou
Supplementary Information accompanies this paper on Cell Death and Disease website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Wei, L., Zhou, Y., Dai, Q. et al. Oroxylin A induces dissociation of hexokinase II from the mitochondria and inhibits glycolysis by SIRT3-mediated deacetylation of cyclophilin D in breast carcinoma. Cell Death Dis 4, e601 (2013). https://doi.org/10.1038/cddis.2013.131
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.131
Keywords
This article is cited by
-
Role of SIRT3 in neurological diseases and rehabilitation training
Metabolic Brain Disease (2023)
-
Mechanism research and treatment progress of NAD pathway related molecules in tumor immune microenvironment
Cancer Cell International (2022)
-
MiR-125b-5p suppresses the bladder cancer progression via targeting HK2 and suppressing PI3K/AKT pathway
Human Cell (2020)
-
Toxoplasma gondii GRA8 induces ATP5A1–SIRT3-mediated mitochondrial metabolic resuscitation: a potential therapy for sepsis
Experimental & Molecular Medicine (2018)
-
Cyclophilin D ablation is associated with increased end-ischemic mitochondrial hexokinase activity
Scientific Reports (2017)