Abstract
During early infection, viruses activate cellular stress-response proteins such as heat-shock proteins (Hsps) to counteract apoptosis, but later on, they modulate these proteins to stimulate apoptosis for efficient viral dissemination. Hsp70 has been attributed to modulate viral entry, transcription, nuclear translocation and virion formation. It also exerts its anti-apoptotic function by binding to apoptosis protease-activating factor 1 (Apaf-1) and disrupting apoptosome formation. Here, we show that influenza A virus can regulate the anti-apoptotic function of Hsp70 through viral protein M1 (matrix 1). M1 itself did not induce apoptosis, but enhanced the effects of apoptotic inducers. M1-small-interfering RNA inhibits virus-induced apoptosis in cells after either virus infection or overexpression of the M1 protein. M1 binds to Hsp70, which results in reduced interaction between Hsp70 and Apaf-1. In a cell-free system, the M1 protein mediates procaspase-9 activation induced by cytochrome c/deoxyadenosine triphosphate. A study involving deletion mutants confirmed the role of the C-terminus substrate-binding domain (EEVD) of Hsp70 and amino acids 128–165 of M1 for this association. The M1 mutants, which did not co-immunoprecipitate with Hsp70, failed to induce apoptosis. Overall, the study confirms the proapoptotic function of the M1 protein during influenza virus infection.
Similar content being viewed by others
Main
Apoptosis or programmed cell death is a genetically and biochemically defined process, which has been implicated in the pathogenesis of various diseases, including bacterial or viral infections.1, 2, 3 During infection, cell death is induced as a consequence of activation of host cellular defense mechanism to limit the virus spread by removing infected cells. In parallel, virus infection or other physiological stress also activates stress responses to facilitate cell survival. For example, activation of anti-apoptotic proteins such as NF-κB, phosphatidylinositol 3-kinase/RAC serine/threonine protein kinase and heat-shock proteins (Hsps) has been observed during early stages of replication to delay apoptosis until completion of their life cycle.4, 5, 6 Conversely, during later stages of infection, viruses exploit the apoptosis machinery for efficient propagation and dissemination by activating signals that either initiate apoptosis or disable anti-apoptotic pathways.
Hsps mainly function as cellular chaperones to maintain protein folding and renaturation.7 Viruses do not have their own chaperone proteins, and thus they rely on cellular chaperones for proper folding of their proteins. There are numerous examples of positive requirement of Hsp40, Hsp70 or Hsp90 by viruses during their replication.8, 9, 10 In contrast, induction of Hsps has also been correlated with inhibition of various RNA viruses, including rotavirus and influenza viruses.11 During physiological stress, other than chaperone activity, Hsps have now been shown to have varied biological functions such as inhibition of protein aggregation, increasing protein disaggregation and modulation of apoptosis by intervening in major apoptotic interactions.12, 13, 14, 15 However, these stress-response functions of Hsps may not always involve their chaperone activity. Many Hsps, especially Hsp70, have been observed to be involved in apoptosis regulation.13, 14, 15 For example, under stress, when Hsp70 levels are high, it has been shown to exert its anti-apoptotic function by binding to apoptosis protease-activating factor 1 (Apaf-1), thereby preventing apoptosome formation and recruitment of caspase-9.14, 15
Influenza A virus, an enveloped RNA virus of the family Orthomyxoviridae, possesses a genome of eight negative-sense RNAs. Influenza viruses have been shown to induce Hsps, of which Hsp90 interacts directly with the polymerase basic 1 (PB1) protein and regulates viral infection,16 whereas Hsp70 has been shown to inhibit influenza virus replication.17 The non-structural protein 1 (NS1) protein of the influenza virus has also been shown to inhibit cleavage of Hsp70 pre-mRNAs to form mature mRNAs.18 In addition, like most eukaryotic viruses, the influenza virus induces apoptosis both in vitro and in vivo.19, 20 To counteract apoptosis during early infection, virus-encoded NS1 protein has been shown to modulate anti-apoptotic proteins.21 However, in a later stage of infection, translocation of the fragment 2 of polymerase basic 1 (PB1-F2) protein to the mitochondrial membrane results in depolarization of membrane potential, release of cytochrome c (cyt c) and activation of caspase-9-mediated intrinsic apoptotic pathway.22, 23 Activation of the mitochondrial apoptotic pathway by interferon-regulatory factor-3-mediated activation of Bax has also been reported during RNA virus infection.24
The M1 of influenza A virus is a phosphorylated 252 amino-acid (aa), major structural protein, which tightly associates with ribonucleoprotein cores while interacting with the membrane envelope and the cytoplasmic tails of spike glycoprotein.25 The M1 protein has been shown to directly bind to heat-shock cognate protein 70 (Hsc70).26 As Hsp70 is the structural homolog of Hsc70 and has also been shown to modulate caspase-9 activation,13, 14, 15, 27, 28 a possible interaction of M1 with Hsp70 was hypothesized. Thus, in this study, we analyzed the possible interaction between M1 and Hsp70 proteins by co-immunoprecipitation experiments. The domains responsible for this interaction were identified in both Hsp70 and M1 using deletion mutants. The study confirms that binding of the M1 protein to Hsp70 disrupts the Hsp70–Apaf-1 complex, resulting in formation of functional apoptosome and activation of caspase-9. Overall, the results emphasize the significant role of virus-encoded proteins in modulating cellular proteins for exploiting the cellular defense machinery to their benefit.
Results
M1 enhances caspase activation mediated by cyt c
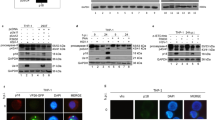
A549 human lung epithelial cells were transiently transfected with either M1-small-interfering RNA (siRNA) (60 nmol) or left untreated for 24 h, followed by infection with influenza APR8 strain (A/Puerto Rico/8/34) at 1 multiplicity of infection (m.o.i.). As a measure of apoptosis, caspase-9 activity was measured by cleavage of a synthetic substrate tetrapeptide LEHD (Leu-Glu-His-Asp) conjugated to 7-amino-4-methyl coumarin (LEHD-AMC). In M1-siRNA-treated and PR8 virus-infected cells, caspase-9 enzymatic activity decreased significantly (≈18-fold) at 14 hour of post infection (h.p.i.) compared with only virus-infected cells (Figure 1a). Concomitantly, caspase-3 activation was also reduced eightfold in M1-siRNA-treated cells (24.6 fluorescence unit) as compared with only virus-infected cells (Figure 1b). Cleavage of caspases into their proteolytically active subunits was further confirmed by immunoblotting in which higher levels of cleavage products of caspase-9 (p37/p35) and caspase-3 (p20/p17) were observed in only PR8 virus-infected cells but not in M1-siRNA- and virus-infected cells (Figure 1c). Effective downregulation of the M1 gene by M1-siRNA was confirmed by immunoblot analysis (Figure 1d). Thus, the results suggest the role of the M1 protein in influenza A virus-induced apoptosis. Activation of caspase-9 during influenza A replication is important as in the presence of caspase-9 and caspase-3 inhibitors, Z-LEHD-FMK (N-benzyloxycarbonyl-LEHD-methyl-fluoromethylketone derivative) and Z-DEVD-FMK (benzyloxycarbonyl-Asp-Glu-Val-Asp-fluoromethylketone), virus replication was significantly reduced in A549 cells (data not shown).
Involvement of M1 in mitochondria-mediated caspase activation during influenza A/PR8 infection. (a and b) A549 cells were transfected with control siRNA or M1 siRNA (60 nmol), and 24 h later, were infected with 1 m.o.i. (multiplicity of infection) of influenza A/PR8 virus. Caspase-9 and caspase-3 activities were determined by hydrolysis of the LEHD-AMC and DEVD-AFC substrates, respectively. Results are representative of three independent experiments. Values represent means±S.D. of one experiment with three measurements taken. (c) Caspase processing was assayed by immunoblot analysis for the indicated times. Zymogens and cleavage products are indicated. (d) Expression of M1 was assessed by immunoblotting in PR8-infected cells and M1-siRNA-treated PR8-infected cells for the indicated times
Although the results suggested the role of the M1 protein during virus-induced apoptosis, it could not be ascertained whether the M1 protein alone is an apoptotic factor or whether it exerts its function in cooperation with other influenza virus proteins. Moreover, M1-siRNA may indirectly result in reduced virus replication, thus affecting caspase-9 activation. To overcome this, the M1 gene was cloned in pcDNA6 (pcD-M1) and transiently expressed in 293T cells. As negative control, cells were transiently transfected with an empty vector (pcDNA6). After 48 h of transfection, cells were treated with staurosporine (1 μM) for 6 h. Activation of caspase-9 was assessed by both fluorometric assay and immunoblotting as described earlier. Staurosporine treatment resulted in activation and cleavage of caspase-9 and caspase-3 in both control and pcD-M1-transfected cells (Figures 2a and b). However, there was four-fold higher activation in pcD-M1-expressing cells as early as 2 h after treatment (Figures 2a and b). When cell extracts were immunoblotted, enhanced cleavage of caspase-9, caspase-7 and caspase-3 was observed in M1-expressing cells than in controls (Figure 2c). Similarly, after staurosporine treatment, cleaved fragment (85kDa) of proapoptotic protein poly (ADP-ribose) polymerase (PARP) was observed early (2 h) in pcD-M1-transfected cells than in empty vector controls (6 h) (Figure 2c). Collectively, these results confirmed the proapoptotic role of the M1 protein.
M1-mediated early activation of caspases in staurosporine induced 293T cells. (a and b) 293T cells were transiently transfected with empty control vector or pcD-M1 and left untreated for 36 h before control and transfected cells were treated with staurosporine (1 μM, 6 h) for the indicated times. Actual enzymatic activity of caspase-9 and caspase-3 in cell lysates of control 293T cells or M1-expressing 293T cells treated with staurosporine was measured fluorometrically by cleavage of LEHD-AMC and DEVD-AFC, respectively, for the given times. Results are representative of three independent experiments. Values represent means±S.D. of one experiment with three measurements taken. (c) Immunoblotting was performed using whole-cell lysates for cleavage of caspase-9, caspase-7, caspase-3 and PARP. β-Actin served as an internal loading control
M1 binds to the Hsp70 protein and disrupts Hsp70–Apaf-1 interaction
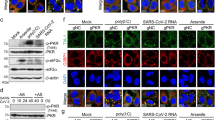
To analyze a possible interaction between Hsp70 and M1, whole-cell lysates after PR8 infection were immunoprecipitated with an anti-Hsp70 polyclonal antibody, followed by immunoblotting with M1 antibody. As shown in Figure 3a, the M1 protein immunoprecipitated with endogenous Hsp70. Furthermore, the co-association was confirmed by reciprocal immunoprecipitation by M1 antibody, followed by immunoblotting with Hsp70 antibody (Figure 3a, lane 3).
M1 association disrupts Hsp70 binding with Apaf-1 during PR8 infection and also in the absence of other viral proteins. (a) Endogenous Hsp70 was immunoprecipitated (IP) from A549 extracts during PR8 infection using an anti-Hsp70 antibody and probed for associated proteins with an anti-M1 antibody (lane 2). In reciprocal immunoprecipitation using an anti-M1 antibody, Hsp70 co-precipitated with M1 (lane 3). (b) Association between Hsp70 and Apaf-1 was analyzed during PR8 infection from 8–26 h.p.i. by immunoprecipitation with an anti-Hsp70 antibody, followed by immunoblotting with an anti-Apaf-1 antibody. In parallel, Hsp70–Apaf-1 association was assessed in cells treated with staurosporine (1 μM). During influenza infection, association between Hsp70 and Apaf-1 was disrupted, whereas Hsp70 immunoprecipitated with Apaf-1 in staurosporine-induced apoptosis. (c) M1-siRNA (50 nmol) was transfected in A549 cells and left untreated for 36 h before PR8 infection. After 14 h.p.i., immunoprecipitation with Apaf-1 antibody was performed to confirm whether M1 binding with Hsp70 disrupted Apaf-1 binding with Hsp70. Result shows that when M1 is downregulated by siRNA, the Apaf-1–Hsp70 association is not affected during PR8 infection. (d) 293T cells were either mock transfected or transfected with the pcD-M1 construct and left untreated for 36 h, followed by staurosporine (1 μM, 6 h) treatment. Cell lysates were then subjected to immunoprecipitation with anti-Hsp70 antibody and immunoblotted with anti-M1 antibody. In parallel, association between Hsp70 with Apaf-1 has been analyzed by immunoprecipitating Hsp70 and probed for Apaf-1. Compared with control staurosporine-treated cells, pcD-M1-transfected cells showed reduced association of Apaf-1 with Hsp70. Immunoglobulin G served as loading control. (e) Further Apaf-1 was immunoprecipitated and probed for Hsp70 to assess its association in pcD-M1 transfected cells
Previous studies have shown direct binding of Hsp70 to Apaf-1, which results in inhibition of its oligomerization and procaspase-9 recruitment.14, 15 To understand whether M1 interaction with Hsp70 affects the Apaf-1–Hsp70 interaction, cells were either treated with staurosporine or infected with the PR8 strain (8, 14, 20 and 26 h) or left untreated. Whole-cell extracts were immunoprecipitated using the anti-Hsp70 antibody, followed by immunoblotting with the Apaf-1 antibody. In untreated and staurosporine-treated cells, Apaf-1 co-immunoprecipitated with Hsp70, whereas weak or no co-immunoprecipitation was observed in PR8 virus-infected cells (Figure 3b). The results confirmed inhibition of the Hsp70–Apaf-1 interaction during influenza virus infection. To assess whether the Hsp70–Apaf-1 dissociation in influenza A virus-infected cells was due to the M1 protein, experiments were conducted in the presence of M1-siRNA. Untransfected or M1-siRNA-transfected cells were infected with the PR8 strain for 14 h, and cell extracts were immunoprecipitated using Apaf-1 antibody. Immunoblotting with the Hsp70 antibody revealed significantly higher co-immunoprecipitation of Hsp70 with Apaf-1 in M1-siRNA-treated cells (Figure 3c), compared with poor association in the absence of M1-siRNA (Figure 3c, lane 3), suggesting that the M1–Hsp70 interaction may result in reduced association of Hsp70 with Apaf-1 during influenza virus infection. Furthermore, to confirm whether M1 inhibits the Hsp70–Apaf-1 interaction directly or indirectly as a part of a multi-protein complex, immunoprecipitation experiments were performed in pcD-M1 or pcDNA-6-transfected 293T cells. After 36 h, cells were treated with staurosporine (1 μM) for 6 h. Whole-cell lysates were immunoprecipitated with the anti-Hsp70 antibody and immunoblotted with anti-M1 and anti-Apaf-1 antibodies. In M1-overexpressing cells, co-immunoprecipitation of Hsp70 and M1 was observed (Figure 3d, middle panel) confirming previous results (Figure 3a), whereas in control cells, significantly higher levels of Apaf-1 co-immunoprecipitated with Hsp70 after staurosporine treatment (Figure 3d). In reciprocal immunoprecipitation using Apaf-1 antibody, Hsp70 co-immunoprecipitated with Apaf-1 in both M1-expressing and control cells (Figure 3e); however, Apaf-1 and Hsp70 interaction was significantly reduced (>3.0-fold) in M1-expressing cells (Figure 3e, lane 2), confirming that the M1 protein can directly bind to Hsp70, which results in reduction in the Hsp70–Apaf-1 complex even in the absence of influenza A virus infection.
M1 inhibits Hsp70-mediated downregulation of cyt c-induced caspase-9 activation
To confirm the mechanism of M1-mediated inhibition of anti-apoptotic function of Hsp70, cyt c-induced activation of caspases was analyzed in an in vitro system using cell-free extracts from Jurkat cells.29 Jurkat cell extracts were used as these cells were shown to have low levels of basal Hsp70 expression.13 Addition of cyt c and deoxyadenosine triphosphate (dATP) resulted in activation of caspase-9 and caspase-3, as determined by cleavage of LEHD-AMC and Asp-Glu-Val-Asp (DEVD) conjugated to 7-amino-4-trifluoromethyl coumarin (DEVD-AFC) substrates (Figures 4a and b). When purified Hsp70 (3.58 μM) protein was added along with cyt c and dATP, significant inhibition of caspase activation was observed (Figures 4a and b). However, when recombinant M1 protein (10 μg/ml) was added along with purified Hsp70, cyt c/dATP-induced caspase activation was restored (Figures 4a and b). Similarly, cleaved 37-kDa (caspase-9) and 19/17-kDa (caspase-3) fragments were observed only in cyt c/dATP and in M1/Hsp70/cyt c/dATP-treated but not in Hsp70/cyt c/dATP-treated cell extracts as assessed by immunoblotting (Figure 4c). These results indicate that M1 interaction suppresses Hsp70-mediated inhibition of the intrinsic pathway during virus infection.
M1 inhibits Hsp70-mediated retardation of caspase activation in a cell-free system. (a and b) Cytosolic extracts prepared from Jurkat T cells were incubated with cytochrome c (10 μM) and dATP (1 mM) in the presence or absence of recombinant human Hsp70 (3.58 μM) and or recombinant M1 (10 μg/ml) for the indicated time periods. Cleavage activities of caspase-9 and caspase-3 were determined by fluorometric quantification of LEHD-AMC and DEVD-AFC substrate cleavage, respectively. Results are representative of three independent experiments. Values represent means±S.D. of one experiment with three measurements taken. (c) Caspase cleavage was assessed by immunoblot analysis after exogenous addition of cytochrome c (10 μM) and dATP (1 mM) in presence or absence of recombinant Hsp70 (3.58 μM) and recombinant M1(10 μg/ml)
Modulation of Hsp70 mRNA and protein levels in PR8 virus-infected cells
If Hsp70 inhibits cellular apoptosis after virus infection, then virus may induce Hsp70 expression for successful completion of its life cycle. mRNA levels of Hsp70 in the nucleus and cytoplasm were measured by real-time PCR at different time points during PR8 virus infection. A significant increase in Hsp70 mRNA in the nucleus (>30-fold at 20 h.p.i.) was observed (Figure 5a), whereas a stable but slight decrease (≈1.5-fold) in Hsp70 mRNA in the cytoplasm was observed (Figure 5a). In parallel, a stable two-fold increase in the heat-shock factor-1 transcript was observed during 8–26 h.p.i. (Figure 5b). As a measure of influenza A virus replication, expression of the M1 gene was analyzed. Significant induction of M1 transcription (20–150-fold) was observed with increasing time after infection (Figure 5c).
Regulation of Hsp70 expression during virus expression. (a) RNA was isolated from nuclear and cytoplasmic fractions of PR8-infected A549 cells using TRIzol reagent. Real-time PCR for the Hsp70 gene was performed using SYBR green reagent. In the nucleus, 25–35-fold increase in Hsp70 mRNA levels was observed at 14–20 h.p.i. In the cytoplasm, 1.5–2-fold decrease in the Hsp70 transcript was observed after PR8 infection (8–26 h.p.i.). (b) In parallel, the HSF1 transcript showed 1.8–2.2-fold increase throughout virus infection. (c) A significant time-dependent increase was observed in mRNA levels of M1 with 80–140-fold increase at 14–26 h.p.i, as assessed by real-time PCR. (d) Hsp70 protein levels during PR8 infection were determined by immunoblotting for indicated time points. Significant increase (3–3.5 fold) was observed at 8–20 h.p.i., followed by downregulation at 26 h.p.i
Increase in Hsp70 protein levels after viral infection of cells has widely been observed.17, 30 Unlike a significant increase in the Hsp70 transcript in the nucleus, only 2.5–3.5-fold increase in the Hsp70 protein was observed after 8–20 h.p.i., followed by downregulation (Figure 5d), which is consistent with the previous report.17, 18 At later time points of infection, basal expression level was observed (data not shown).
M1 binds to the SBD of Hsp70
The C-terminal region of Hsp70 has a regulatory EEVD motif, which has been shown to be crucial for its activity. Deletion (Hsp70ΔEEVD) or substitution with alanine residues for the four C-terminal aa (Hsp70AAAA) disrupts its intramolecular regulation and intermolecular interactions.31 The pFLAG-CMV6-M1 construct was co-transfected with either pcD-Hsp70 or the Hsp70 mutants (pcD-Hsp70ΔEEVD or pcD-Hsp70AAAA) in 293T cells. After immunoprecipitation of whole-cell extracts using anti-Flag antibody, only native Hsp70 co-precipitated (Figure 6a, lane 2). Mutants Hsp70AAAA and Hsp70 ΔEEVD were unable to bind M1 in the cellular environment (Figure 6a). For confirming direct interaction of proteins under cell-free conditions, native Hsp70, Hsp70AAAA and Hsp70ΔEEVD proteins were immobilized on nickel beads and then incubated either with the M1 protein or mock control. After eluting the bead complexes, immunoblotting with anti-M1 antibody showed that M1 associated with native Hsp70 but not with Hsp70AAAA or Hsp70ΔEEVD proteins in pull-down experiments (Figure 6b). These results confirmed that the C-terminal EEVD motif of the substrate-binding domain (SBD) region of Hsp70 is critical for its interaction with M1.
The EEVD motif of Hsp70 is responsible for binding to 128–165 aa of M1. (a) pFLAG-CMV6-M1 was co-transfected with pcD-Hsp70, pcD-Hsp70AAAA or pcD-Hsp70ΔEEVD in 293T cells to assess the importance of EEVD motif in in vivo binding. Immunoprecipitation was performed with anti-Flag antibody to pull down the Flag-M1 protein. Immunoblotting showed that only native Hsp70 was precipitated along with M1 but not with the mutants. (b) Importance of the EEVD motif in M1 binding was confirmed by an in vitro binding assay. Purified His-tagged Hsp70, Hsp70AAAA or Hsp70ΔEEVD proteins (10 μg) were pre-immobilized on Ni+2 in HBS buffer overnight at 4 °C. After washing, beads were incubated with recombinant M1 protein (5 μg), then washed and proteins were immunoblotted with anti-M1 antibody. Only native Hsp70 directly bound with the M1 protein. (c) Schematic drawing of M1 deletion mutants used in this study. Deleted amino acids are indicated by dotted lines. (d) Immunodetection of Hsp70 after Flag-M1 immunoprecipitation of 293T cells transfected with an empty vector (control), or with a vector containing either wild-type M1 cDNA or the mutants described in panel c (left panel). Expression of wild-type and mutants of M1 protein in 293T cells was confirmed by immunoblotting
C-terminal motif of M1 interacts with Hsp70 leading to caspase-9 activation
Involvement of the C-terminal region of Hsp70 in M1 binding was confirmed; thus, to identify the region of the M1 protein involved in this binding, deletion mutants of M1 were constructed (Figure 6c) based on a previous report.26 293T cells were transiently transfected with an empty vector or pFLAG-M1 or mutants of M1 (Figure 6d). Co-immunoprecipitation experiments demonstrated that deletion mutants Δ128–165 and Δ128–201 did not bind to Hsp70, whereas mutants Δ101–127, Δ101–150 and Δ166–201 co-associated with Hsp70 as efficiently as did wild-type M1 (Figure 6d). All M1 mutants that failed to immunoprecipitate Hsp70 also lost their ability to induce staurosporine-mediated caspase-9 and caspase-3 activation (Table 1). Purified M1, M1Δ101–127 and M1Δ128–165 proteins were immobilized on nickel beads and incubated with Hsp70. Results indicated that unlike wild-type M1 protein and mutant M1Δ101–127 protein, mutant M1Δ128–165 protein did not bind to Hsp70 protein (Figure 7a). Furthermore, in Hsp70-immunodepleted Jurkat cell extracts, Apaf-1 co-immunoprecipitated with the Hsp70 protein in the presence of mutant M1Δ128–165, whereas wild-type M1 disrupted this binding (Figure 7b). In addition, in the presence of mutant Δ128–165 protein, no activation of caspase-9 was observed in cell-free extracts supplemented with cyt c/dATP and Hsp70 (Figure 7c). Collectively, these results confirm the role of the C-terminal region (aa 128–165) of the M1 protein in Hsp70 binding and induction of apoptosis.
Amino acid 128–165 is responsible for direct binding with Hsp70 to prevent Apaf-1 binding and to aid in caspase-9 processing induced by cytochrome c/ATP. (a) His-tagged M1or M1Δ128–165 (10 μg) was immobilized on Ni+2 overnight in HBS at 4 °C, and then washed and incubated with recombinant Hsp70 (10 μg) for 4 h at 4 °C. Associated Hsp70 was detected by immunoblotting. Wild-type Hsp70 associated with native M1 but not with M1Δ128–165. (b) Jurkat cell extracts were Hsp70 immunodepleted, before recombinant Hsp70 and M1 or M1Δ128–165 proteins were added to it incubated and Apaf-1 was immunoprecipitated. Hsp70 did not associate with Apaf-1 when native M1 was present, but did associate in the absence of M1 or in the presence of M1Δ128–165 protein. (c) Co-addition of recombinant M1 enhances processing of procaspase-9 induced by dATP or ATP and cytochrome c in Jurkat cell extracts in the presence of wild-type Hsp70, whereas in the presence of M1Δ128–165 protein, caspase-9 activation is inhibited
Discussion
Viruses exploit the existing cellular machinery for regulating their own gene expression. Viral genes are translated based on their requirements during different stages of the viral life cycle. For example, during influenza virus infection, synthesis of nucleoprotein and NS1 protein is favored during early infection, whereas synthesis of HA, NA, PB1-F2 and M1 genes is delayed.32 NS1 has been shown to prevent apoptosis and interferon induction during early stages,21 whereas PB1-F2 disrupts mitochondrial function resulting in release of cyt c and apoptosis for efficient dissemination of viral progeny during later stages of infection.22, 23 Concurrently, a co-relation between activation of caspase-9 and expression of M1 was also observed at 14 h.p.i. (Figures 1c and d). The proapoptotic role of M1 was validated, as virus-induced activation of caspase-9 and caspase-3 was inhibited in the presence of M1-siRNA, and in cells expressing the M1 protein, apoptotic inducers like staurosporine induced significantly higher apoptosis (Figure 2).
The functional role of Hsp chaperone proteins in virus entry, replication, transcription, virion assembly, translational regulation, transformation and mainly protein folding and maturation during infection is well documented.10, 33 Increased expression of the Hsp70 gene during viral infection has also been reported.30 During this study, we observed a time-dependent increase (25–40-fold) in Hsp70 mRNA in the nucleus but not in the cytoplasm (Figure 5a). It can be postulated that NS1 protein's ability to inhibit translocation of pre-mRNAs to the cytoplasm18 may have resulted in accumulation of Hsp70 transcripts in the nucleus. Furthermore, only 2.5–3.5-fold upregulation of the Hsp70 protein was observed at 8–20 h.p.i., followed by decrease at later time points (Figure 5d), which is consistent with the previous report.17 In spite of low levels of transcripts in the cytoplasm, increased but steady levels of the Hsp70 protein in cells could be due to increased stability of the Hsp70 protein and transcripts during cellular stress to maintain cellular homeostasis.34, 35 However, levels of the Hsp70 protein in cells after virus infection are also regulated stringently, as virus production was shown to be inhibited in heat-induced cells.10
On the basis of previous reports and our results, four observations were noted, namely (1) influenza A virus induces stress proteins including Hsp70 in cells after infection30 (Figures 5a and d), (2) under stress when Hsp70 is upregulated, it can inhibit apoptosis by binding to Apaf-1, which results in inhibition of apoptosome formation and caspase-9 recruitment, although basal levels of Hsp70 are essential during assembly of functional apoptosome,13, 14, 15 (3) during influenza A virus infection, the M1 protein modulates activation of caspase-9 and cyt c-mediated intrinsic pathway (Figures 1a–c, Supplementary Figure S1) and (4) the M1 protein binds to Hsc70, a homolog of the Hsp70 protein.26 Thus, we hypothesized that M1 may bind to Hsp70 to modulate its anti-apoptotic function, thus resulting in enhanced caspase-9 activation. Co-immunoprecipitation experiments showed that the M1 protein expressed either through virus infection or that the expression vector binds to Hsp70 (Figure 3, Supplementary Figure S2). In the presence of the M1 protein, Hsp70–Apaf-1 interaction was significantly reduced. Consistent with caspase activation results (Figures 1a–c), in the presence of M1-siRNA, Hsp70–Apaf-1 binding was restored (Figure 3c). In the cell-free system also, purified M1 protein restored activation of caspase-9 in the presence of Hsp70, cyt c and dATP (Figures 4a–c), further confirming that direct interaction between Hsp70 and M1 is responsible for inhibition of the anti-apoptotic function of Hsp70. These results are consistent with previous reports, which propose that at higher concentrations, Hsp70 is anti-apoptotic either because of the formation of high molecular-weight aggregates of Hsp70/Apaf-1 or because of the change in conformation of apoptosome which prevents recruitment of caspase-9.13, 14, 15 On the basis of our results, it can be hypothesized that virus-encoded protein M1 quenches excess Hsp70, which is induced in the cells as a consequence of virus infection, so that Apaf-1-mediated apoptosome could be formed for initiation of apoptosis. Apoptosis is required by viruses for their proper dissemination, which is the case for influenza A virus also, as in the presence of caspase-9 and caspase-3 inhibitors, virus replication and infection were inhibited (data not shown), which is consistent with previous reports.36 Low levels of Apaf-1–Hsp70 association were still observed during PR8-induced apoptosis, as physiological levels of Hsp70 have been shown to be essential for Apaf-1- and cyt c-mediated functional apoptosome formation.14
Deletion mutants of the Hsp70 gene (Figures 6a and b) confirmed significance of the C-terminus acidic region of Hsp70 for interaction with M1, which could bind only to full-length Hsp70 protein, but not to Hsp70AAAA or Hsp70EEVD. The extreme C-terminal-regulatory EEVD motif of Hsp70, essential for its ATPase activity and substrate binding is highly conserved within the Hsp70 family.31
The RNA-binding domain (aa 76–103) of the M1 protein was not found to be essential, but aa 102–201 was shown to be necessary for M1-Hsc70 binding.26 Using this as a reference, several deletion mutants of M1 were constructed spanning the aa 101–201 region. In addition, Hsp70-binding sites on M1 were also predicted using LIMBO software (http://limbo.vub.ac.be/), an algorithm for identifying chaperone-binding sites in proteins. This algorithm uses data from binding of bacterial homolog of Hsp70 (DnaK) to cellulose-immobilized peptides and also from a sequence-based profile or a position-specific scoring matrix.37 The binding motif for Hsp70 consists of a heptamer having a hydrophobic core of 4–5 residues enriched particularly in Leucine, Isoleucine, Valine, Phenylalanine, tyrosine and two flanking basic residues that complement the overall negatively charged DnaK surface.38 Four most probable DnaK-binding motifs were predicted, and out of which except aa 128–134, other three are excluded by our deletion mutation experiments. As shown in Figure 6d, 128–201 and 128–165 mutants of M1 failed to immunoprecipitate Hsp70. Mutants preventing M1–Hsp70 interaction failed to activate caspase-9 (Figure 7c), further suggesting that this interaction is required for the apoptotic effect of M1 during infection. Moreover, as Hsp70 binds to the cellular caspase recruitment domain (CARD) domain of Apaf-1,15 it is expected to have one or more common functional domains between the CARD motif and aa 128–165 region of the M1 protein. The eukaryotic linear motif (http://elm.eu.org/) program predicted occurrence of two common domain-binding motifs viz. Forkhead-associated domain 1-binding motif that spans 137–143 aa and PSD-95, Dlg, ZO-1/2 domain 3 (PSD-95, Dlg, ZO-1/2 domain 3)-binding motif spanning 151–154 aa between the two proteins, suggesting both Apaf-1 and M1 as binding partners of Hsp70.
In addition to caspase-9, Hsp70 has been reported to intervene with other apoptotic signaling cascades, such as caspase-8, c-Jun N-terminal kinase (JNK/SAPK) and apoptosis signal-regulating kinase 1 activation.39 As caspase-8 is activated during influenza A infection, its intervention may prevent cleavage of BH3-interacting domain death agonist (Bid) into tBid (truncated BH3-interacting domain death agonist), thus inhibiting amplification of the intrinsic pathway (Supplementary References). However, in the presence of caspase-8 inhibitor, PR8 infection still activated caspase-9, suggesting it to be independent of caspase-8 activation (Supplementary Figure S3). Thus, even though the results of this study focused on M1-mediated activation of caspase-9 due to reduction in Hsp70–Apaf-1 interaction and resulting in activation of apoptosis, suppression of anti-apoptotic activity of Hsp70 by M1 can be attributed to its overall apoptotic function. Moreover, in the presence of M1-siRNA, cleavage of bid and phosphorylation of JNK/SAPK was inhibited (Supplementary Figure S4). Overall, the results assert the significant role of virus-encoded proteins in regulation of diverse host stress-response pathways during virus infection.
Materials and Methods
Viruses, cells and viral infection
Madin-Darby canine kidney (MDCK), human alveolar epithelial cells (A549), 293T and Jurkat cells were grown in MEM, Ham's F12, DMEM and RPMI 1640 media supplemented with 10% heat-inactivated fetal bovine serum, 2 mM L-glutamine, 2 mM sodium pyruvate and 1 × PSF (penicillin, streptomycin and fungizone), respectively, at 37 °C with 5% CO2. For infection, cells were washed with phosphate-buffered saline (PBS) and infected with PR8 at the indicated m.o.i. in PBS/BA (PBS containing 0.2% bovine serum albumin (BSA), 1 mM MgCl2, 0.9 mM CaCl2, 100 Units/ml penicillin and 0.1 mg/ml streptomycin) for 45 min at 37 °C. The inoculum was aspirated and A549 or MDCK cells were incubated in respective media supplemented with 0.2% BSA and antibiotics. The amount of infectious virus in cell supernatants was determined by plaque assay as described previously.40
Construction of vectors
Vectors were constructed using the primers having restriction sites as follows:

Antibodies, reagents and inhibitors
Antibodies against M1 (Sc-69824, Sc-17589) and cyt c (Sc-13560) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). β-Actin (551527), Apaf-1 (611365), caspase-8 (551244) and Bax (610983)-specific antibodies were obtained from BD Biosciences (San Diego, CA, USA). Antibodies against caspase-9 (9501, 9502), caspase-7 (9491, 9491), caspase-3 (9662, 9664), PARP (9541, 9542), Bid (2002) and Flag (DYKDDDDK, 2368) were from Cell Signaling Technology, Inc (Danvers, MA, USA). Hsp70 antibody was obtained from Upstate (Billerica, MA, USA) (NBPI-49250) and BD Biosciences (554243). All antibodies were used at 1 : 1000 dilution, except anti-M1 and anti-β-actin, which were used at 1 : 500. Staurosporine (Sigma, St. Louis, MO, USA; S5921) was used at 1 μM/ml, whereas Horse-heart cyt c (Sigma, C7752) was used at 10 μM/ml. ATP (NEB, P0756S) or dATP (NEB, N0440S) was used at 1 mM/ml. Z-IETD-FMK (BD, 550380) was used at 80 μM/ml.
Plasmid and siRNA transfection
293T, A549 and Jurkat cells were either transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) or siPORT-NeoFX (Ambion, Austin, TX, USA) according to the manufacturer's instructions. Custom-synthetic siRNA (5′-CTCCAGATTTGCCTGAAGA-3′) against M1 was obtained from Dharmacon (Lafayette, CO, USA). Control siRNA was obtained from Qiagen (Hilden, Germany) (All Star Negative Control, 1027280).
Western blot analysis
Total protein was extracted with Totex buffer (20 mM Hepes at pH 7.9, 0.35 M NaCl, 20% glycerol, 1% NP-40, 1 mM MgCl2, 0.5 mM EDTA, 0.1 mM EGTA, 50 mM NaF and 0.3 mM NaVO3) containing a mixture of protease and phosphatase inhibitors (Sigma). Immunoblotting was performed with specific antibodies and visualized using ECL western blotting detection kit (Millipore, Billerica, MA, USA).
Cell fractionation
Cytosolic extracts free of the nuclei and mitochondria were prepared. In brief, cells were washed in ice-cold PBS, pH 7.2, and then in hypotonic extraction buffer (HEB: 50 mM PIPES pH 7.4,50 mM KCl, 5 mM EGTA, 2 mM MgCl2, 1 mM dithiothreitol and 0.1 mM phenylmethylsulfonyl fluoride (PMSF)) and centrifuged. The pellet was resuspended in HEB and lysed in a Dounce homogenizer. This cell lysate was centrifuged for 30 min at 16 000 × g at 4 °C, and the clarified supernatant was either tested immediately or stored in aliquots at −80 °C. Mitochondrial fractions were prepared by resuspending cells in ice-cold buffer A (250 mM sucrose, 20 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 17 mg/ml PMSF, 8 mg/ml aprotinin, 2 mg/ml leupeptin (pH 7.4)) and homogenized in a Potter-Thomas homogenizer. Nuclei were pelleted by a 10-min 750 g spin. The supernatant was spun at 10 000 × g for 25 min and the mitochondrial fraction resuspended in buffer A and centrifuged at 10 000 × g for 25 min. Mitochondrial pellets were resuspended and kept in buffer (4 °C) containing 7 M urea, 2 M thiourea, 4% CHAPS, 120 mM dithiothreitol (DTT), 2% ampholytes (pH 3–10) and 40 mM Tris-HCl for 30 min.
Co-immunoprecipitation
Cells were washed with ice-cold PBS and then lysed in a solution containing 10 mM Tris (pH 8.0), 170 mM NaCl, 0.5% NP-40 and protease inhibitors for 30 min on ice. Cell debris were removed by centrifugation, and the supernatants were incubated with anti-Flag or M1 or Apaf-1 and anti-Hsp70 antibodies overnight at 4 °C and with protein A-Sepharose for a further 4 h. Beads were washed four times with 1 ml of wash buffer (200 mM Tris at pH 8.0, 100 mM NaCl and 0.5% NP-40). Bound proteins were eluted with SDS sample buffer and separated on 12% SDS-PAGE gels before immunoblotting with anti-Hsp70 and M1 or Apaf-1 antibody, respectively.
Quantitative real-time PCR
Total RNA was isolated using TRIzol (Invitrogen) according to the manufacturer's instructions. Nuclear and cytoplasmic RNA were isolated from previously fractionated nuclear and cytoplasmic extracts of cells at 4 °C using TRIzol. cDNA was prepared from 1–2 μg of RNA using the Superscript III reverse transcriptase (Invitrogen) with random hexamer primers. Real-time PCR reactions (50 °C for 2 min, 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s and 72 °C for 10 min) were performed in triplicate using SYBR Green (Applied Biosystems, Foster City, CA, USA). Primer sequences are available on request.
Protein expression and purification
Proteins were expressed in C41 (DE3) strain Escherichia coli cells, grown at 37 °C to an OD600 of 1.0, and then induced with 0.5 or 1.0 mM isopropylthiogalactoside and incubated further at 4 h. The pelleted cells were lysed in buffer A (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris-Cl, pH 8.0) and centrifuged. The resin was washed with buffer A, and the lysed supernatant was passed through Ni-NTA spin column (Qiagen). The Ni-NTA spin column was washed with buffer B (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris-Cl, pH 6.3) and finally the bound protein was eluted with elution buffer (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris-Cl, pH 4.5). The eluted protein was then refolded to its native conformation by dialysis for in vitro binding assays. Dialysis buffer contained urea (8, 6, 4, 2, 1 and 0 M), 5 mM reduced glutathione, 0.5 mM oxidized glutathione, 50 mM glycine; pH 9.0, 5 mM EDTA, 10% glycerol, 10% glucose, 10% sucrose, 250 mM NaCl, 2 mM MgCl2, 50 mM HEPES-KOH (pH 7.5–7.9), 0.5 mM PMSF, 2 μg/ml aprotinin and 5 μg/ml leupeptin. Proteins were stored in aliquots at −80 °C. Whenever necessary, purified proteins were subjected to enterokinase cleavage (EK: substrate ratio=1 : 42 at 37 °C in 50 mM Tris, pH 7.6; reaction was stopped by adding 1 mM PMSF and heating at 95 °C for 10 min and then passed through the Ni-NTA column) to remove His tag.
Preparation of cell-free extracts and in vitro caspase activation
Jurkat cells were collected by centrifugation and washed twice in ice-cold PBS. The cell pellet was resuspended in an equal volume of HEB, incubated on ice for 15 min and lysed by 10-s vigorous vortexing after addition of 5% NP-40. Lysates were then centrifuged for 5 min at 15 000 × g, and the cytoplasmic supernatant was stored at −70 °C. Endogenous caspase-9 was activated by adding 1 mM dATP/ATP (NEB) and 10 μM horse-heart cyt c (Sigma) to cytosolic extracts (300 μg protein equivalent) and incubating at 37 °C for respective times. Reactions were stopped by adding 5 × SDS loading buffer and boiling for 5 min. Aliquots (50 μg) were also removed for assessment of caspase processing by immunoblot analysis.
Assay for caspase-9 and caspase-3 activation
Cell lysates from control cells and M1-siRNA-treated cells infected with PR8, pcD-M1-transfected cell or extracts of in vitro caspase activation assay, were prepared as described previously. Actual caspase-9 and caspase-3 cleavage activity was measured using ApoAlert caspase fluorescent assay kit according to the manufacturer's instructions (Clontech, Mountain View, CA, USA) that uses synthetic substrates LEHD-AMC and DEVD-AFC, respectively.
In vitro association of recombinant M1 with Hsp70
Recombinant native M1 and the M1 mutants (∼5 μg) were previously immobilized on Ni2+ by overnight incubation in HEPES-buffered saline (HBS) at 4 °C. After extensive washing in PBS/0.3% Tween to remove unbound protein, M1 proteins were incubated with Hsp70 (10 μg) or Hsp70AAAA (10 μg) for 4 h at 4 °C in HBS. Beads were washed extensively with HBS to remove non-specifically bound proteins. Remaining proteins were separated by 4 × sample buffer and then analyzed by SDS-PAGE and immunoblotted for Hsp70. Similarly, in reciprocal experiments, recombinant Hsp70 or Hsp70AAAA or Hsp70EEVD (∼5 μg) were pre-immobilized on Ni2+, followed by binding and immunoblotting of M1 (10 μg) protein.
In vitro procaspase-9 activation in the presence of recombinant Hsp70, M1 and mutants
Cyt c (10 μg) and dATP/ATP (1 mM) were incubated in the presence or absence of Hsp70 (5 μg) or Hsp70AAAA (5 μg) or Hsp70 (5 μg) and M1 (10 μg) or Hsp70 (5 μg) and M1Δ128–165(10 μg) in cytosolic Jurkat cell extracts (50 μg protein equivalent) at 37 °C. Hsp70 and M1 proteins were pre-incubated for 30 min at 5 °C before dATP/ATP and cyt c addition. Procaspase-9 cleavage was then assessed by immunoblotting as described previously.
Statistical analysis
Data are expressed as mean±S.D. of at least three independent experiments (n≥3). In all tests, P 0.05 was considered statistically significant.
Abbreviations
- Apaf-1:
-
apoptosis protease-activating factor 1
- M1:
-
matrix protein 1
- siRNA:
-
small-interfering RNA
- dATP:
-
deoxyadenosine triphosphate
- Hsp:
-
heat-shock protein
- PB1:
-
polymerase basic 1
- NS1:
-
non-structural protein 1
- PB1-F2:
-
fragment 2 of polymerase basic 1
- Hsc70:
-
heat-shock cognate protein 70
- LEHD-AMC:
-
tetrapeptide LEHD (Leu-Glu-His-Asp) conjugated to 7-amino-4-methyl coumarin
- Z-LEHD-FMK:
-
N-benzyloxycarbonyl-LEHD-methyl-fluoromethylketone derivative
- Z-DEVD-FMK:
-
benzyloxycarbonyl-Asp-Glu-Val-Asp-fluoromethylketone
- PARP:
-
poly (ADP-ribose) polymerase
- cyt c:
-
cytochrome c
- DEVD-AFC:
-
Asp-Glu-Val-Asp (DEVD) conjugated to 7-amino-4-trifluoromethyl coumarin
- ∣SBD:
-
substrate-binding domain
- h.p.i.:
-
hour of post infection
- PR8:
-
8th strain of influenza A virus isolated from Puerto Rico
- DnaK:
-
bacterial homolog of Hsp70
- CARD:
-
caspase recruitment domain
- JNK/SAPK:
-
c-Jun N-terminal kinase
- apoptosis signal-regulating kinase 1∣Bid:
-
BH3-interacting domain death agonist
- tBid:
-
truncated BH3-interacting domain death agonist
References
Barber GN . Host defense, viruses and apoptosis. Cell Death Differ 2001; 8: 113–126.
Hardwick JM . Apoptosis in viral pathogenesis. Cell Death Differ 2001; 8: 109–110.
Lamkanfi M, Dixit VM . Manipulation of host cell death pathways during microbial infections. Cell Host Microbe 2010; 8: 44–54.
Benedict CA, Norris PS, Ware CF . To kill or be killed: viral evasion of apoptosis. Nat Immunol 2002; 3: 1013–1018.
Bagchi P, Dutta D, Chattopadhyay S, Mukherjee A, Halder UC, Sarkar S et al. Rotavirus nonstructural protein 1 suppresses virus-induced cellular apoptosis to facilitate viral growth by activating the cell survival pathways during early stages of infection. J Virol 2010; 84: 6834–6845.
Ehrhardt C, Ludwig S . A new player in a deadly game: influenza viruses and the PI3K/Akt signalling pathway. Cell Microbiol 2009; 11: 863–871.
Hartl FU, Hayer-Hartl M . Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol 2009; 16: 574–581.
Dutta D, Bagchi P, Chatterjee A, Nayak MK, Mukherjee A, Chattopadhyay S et al. The molecular chaperone heat shock protein-90 positively regulates rotavirus infection. Virology 2009; 391: 325–333.
Glotzer JB, Saltik M, Chiocca S, Michou AI, Moseley P, Cotton M . Activation of heat-shock response by an adenovirus is essential for virus replication. Nature 2000; 407: 207–211.
Lin BY, Makhov AM, Griffith JD, Broker TR, Chow LT . Chaperone proteins abrogate inhibition of the human papillomavirus (HPV) E1 replicative helicase by the HPV E2 protein. Mol Cell Biol 2002; 22: 6592–6604.
Broquet AH, Lenoir C, Gardet A, Sapin C, Chwetzoff S, Jouniaux AM et al. HSP70 negatively controls rotavirus protein bioavailability in Caco-2 cells infected by rotavirus RF strain. J Virol 2007; 81: 1297–1304.
Ben-Zvi A, De Los Rios P, Dietler G, Goloubinoff P . Active solubilization and refolding of stable protein aggregates by cooperative unfolding action of individual hsp70 chaperones. J Biol Chem 2004; 279: 37298–37303.
Beere HM, Wolf BB, Cain K, Mosser DD, Mahboubi A, Kuwana T et al. Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat Cell Biol 2000; 2: 469–475.
Kim HE, Jiang X, Du F, Wang X . PHAPI, CAS, and Hsp70 promote apoptosome formation by preventing Apaf-1 aggregation and enhancing nucleotide exchange on Apaf-1. Mol Cell 2008; 30: 239–247.
Saleh A, Srinivasula SM, Balkir L, Robbins PD, Alnemri ES . Negative regulation of the Apaf-1 apoptosome by Hsp70. Nat Cell Biol 2000; 2: 476–483.
Naito T, Momose F, Kawaguchi A, Nagata K . Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J Virol 2007; 81: 1339–1349.
Hirayama E, Atagi H, Hiraki A, Kim J . Heat shock protein 70 is related to thermal inhibition of nuclear export of the influenza virus ribonucleoprotein complex. J Virol 2004; 78: 1263–1270.
Shimizu K, Iguchi A, Gomyou R, Ono Y . Influenza virus inhibits cleavage of the HSP70 pre-mRNAs at the polyadenylation site. Virology 1999; 254: 213–219.
Mori I, Komatsu T, Takeuchi K, Nakakuki K, Sudo M, Kimura Y . In vivo induction of apoptosis by influenza virus. J Gen Virol 1995; 76: 2869–2873.
Takizawa T, Matsukawa S, Higuchi Y, Nakamura S, Nakanishi Y, Fukuda R . Induction of programmed cell death (apoptosis) by influenza virus infection in tissue culture cells. J Gen Virol 1993; 74: 2347–2355.
Shin YK, Liu Q, Tikoo SK, Babiuk LA, Zhou Y . Influenza A virus NS1 protein activates the phosphatidylinositol 3-kinase (PI3K)/Akt pathway by direct interaction with the p85 subunit of PI3K. J Gen Virol 2007; 88: 13–18.
Henkel M, Mitzner D, Henklein P, Meyer-Almes FJ, Moroni A, Difrancesco ML et al. The proapoptotic influenza A virus protein PB1-F2 forms a nonselective ion channel. PLoS One 2010; 5: e11112.
Zamarin D, García-Sastre A, Xiao X, Wang R, Palese P . Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog 2005; 1: e4.
Chattopadhyay S, Marques JT, Yamashita M, Peters KL, Smith K, Desai A et al. Viral apoptosis is induced by IRF-3-mediated activation of Bax. EMBO J 2010; 29: 1762–1773.
Harris A, Forouhar F, Qiu S, Sha B, Luo M . The crystal structure of the influenza matrix protein M1 at neutral pH: M1-M1 protein interfaces can rotate in the oligomeric structures of M1. Virology 2001; 289: 34–44.
Watanabe K, Fuse T, Asano I, Tsukahara F, Maru Y, Nagata K . Identification of Hsc70 as an influenza virus matrix protein (M1) binding factor involved in the virus life cycle. FEBS Lett 2006; 580: 5785–5790.
Brehmer D, Rüdiger S, Gässler CS, Klostermeier D, Packschies L, Reinstein J . Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat Struct Biol 2001; 8: 427–432.
Mayer MP, Bukau B . Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 2005; 62: 670–684.
Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 1997; 91: 479–489.
LaThangue NB, Shriver K, Dawson C, Chan WL . Herpes simplex virus infection causes the accumulation of a heat-shock protein. EMBO J 1984; 3: 267–277.
Freeman BC, Myers MP, Schumacher R, Morimoto RI . Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J 1995; 14: 2281–2292.
Hatada E, Hasegawa M, Mukaigawa J, Shimizu K, Fukuda R . Control of influenza virus gene expression: quantitative analysis of each viral RNA species in infected cells. J Biochem (Tokyo) 1989; 105: 537–546.
Mayer MP . Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Rev Physiol Biochem Pharmacol 2005; 153: 1–46.
Concannon CG, FitzGerald U, Holmberg CI, Szegezdi E, Sistonen L, Samali A . CD95-mediated alteration in Hsp70 levels is dependent on protein stabilization. Cell Stress Chaperones 2005; 10: 59–65.
Theodorakis NG, Morimoto RI . Posttranscriptional regulation of hsp70 expression in human cells: effects of heat shock, inhibition of protein synthesis, and adenovirus infection on translation and mRNA stability adenovirus infection on translation and mRNA stability. Mol Cell Biol 1987; 7: 4357–4368.
Wurzer WJ, Planz O, Ehrhardt C, Giner M, Silberzahn T, Pleschka S et al. Caspase 3 activation is essential for efficient influenza virus propagation. EMBO J 2003; 22: 2717–2728.
Van Durme J, Maurer-Stroh S, Gallardo R, Wilkinson H, Rousseau F, Schymkowitz J . Accurate prediction of DnaK-peptide binding via homology modelling and experimental data. PLoS Comput Biol 2009; 5: e1000475.
Rudiger S, Schneider-Mergener J, Bukau B . Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J 2001; 20: 1042–1050.
Gabai VL, Mabuchi K, Mosser DD, Sherman MY . Hsp72 and stress kinase c-jun N-terminal kinase regulate the bid-dependent pathway in tumor necrosis factor-induced apoptosis. Mol Cell Biol 2002; 22: 3415–3424.
Ludwig S, Ehrhardt C, Neumeier ER, Kracht M, Rapp UR, Pleschka S . Influenza virus-induced AP-1-dependent gene expression requires activation of the JNK signaling pathway. J Biol Chem 2001; 276: 10990–10998.
Acknowledgements
This study was partially supported by the Indian Council of Medical Research (ICMR), India. We sincerely thank lab members Dr. AS Agarwal, T Roy, R Bhowmik, A Mukherjee and S Nandi for their support and encouragement in the lab. We are thankful to Prof. Alexandra Newton (University of California at San Diego) for Hsp70 construct. UCH and PB are supported by the Senior Research Fellowship from the Council of Scientific and Industrial Research (CSIR), India. DD is supported by the University Grants Commission (UGC), India.
Author Contributions. UCH designed and performed the experiments. UCH and MC-S contributed in conceiving and writing the manuscript. PB, SC and DD contributed in cloning of various constructs and cell culture and infection during the study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Stephanou
Supplementary Information accompanies the paper on Cell Death and Disease website
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Halder, U., Bagchi, P., Chattopadhyay, S. et al. Cell death regulation during influenza A virus infection by matrix (M1) protein: a model of viral control over the cellular survival pathway. Cell Death Dis 2, e197 (2011). https://doi.org/10.1038/cddis.2011.75
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2011.75
Keywords
This article is cited by
-
Influenza viral matrix 1 protein aggravates viral pathogenicity by inducing TLR4-mediated reactive oxygen species production and apoptotic cell death
Cell Death & Disease (2023)
-
Influenza A virus-induced apoptosis and virus propagation
Apoptosis (2020)
-
Nucleoprotein of influenza A virus negatively impacts antiapoptotic protein API5 to enhance E2F1-dependent apoptosis and virus replication
Cell Death & Disease (2015)
-
Interplay between influenza A virus and host factors: targets for antiviral intervention
Archives of Virology (2015)
-
Influenza A virus nucleoprotein induces apoptosis in human airway epithelial cells: implications of a novel interaction between nucleoprotein and host protein Clusterin
Cell Death & Disease (2013)